Sodium »
PDB 9jfb-9mbf »
9kyo »
Sodium in PDB 9kyo: Ges Bound Mtaut
Other elements in 9kyo:
The structure of Ges Bound Mtaut also contains other interesting chemical elements:
| Chlorine | (Cl) | 1 atom |
Sodium Binding Sites:
The binding sites of Sodium atom in the Ges Bound Mtaut
(pdb code 9kyo). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 2 binding sites of Sodium where determined in the Ges Bound Mtaut, PDB code: 9kyo:
Jump to Sodium binding site number: 1; 2;
In total 2 binding sites of Sodium where determined in the Ges Bound Mtaut, PDB code: 9kyo:
Jump to Sodium binding site number: 1; 2;
Sodium binding site 1 out of 2 in 9kyo
Go back to
Sodium binding site 1 out
of 2 in the Ges Bound Mtaut
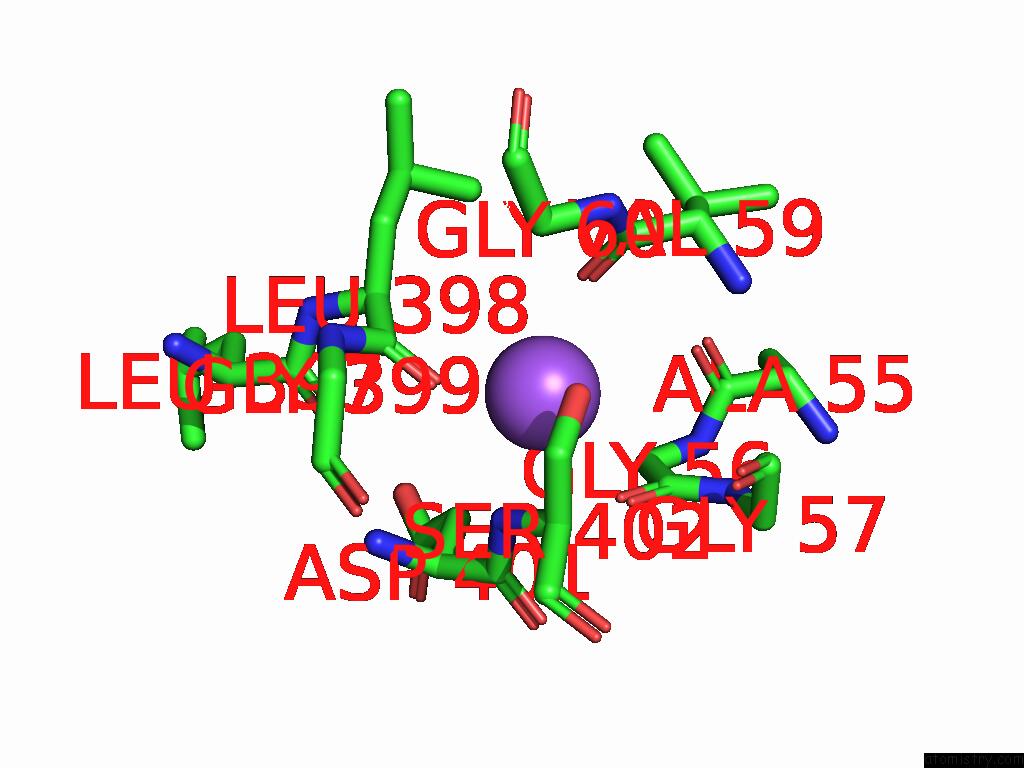
Mono view
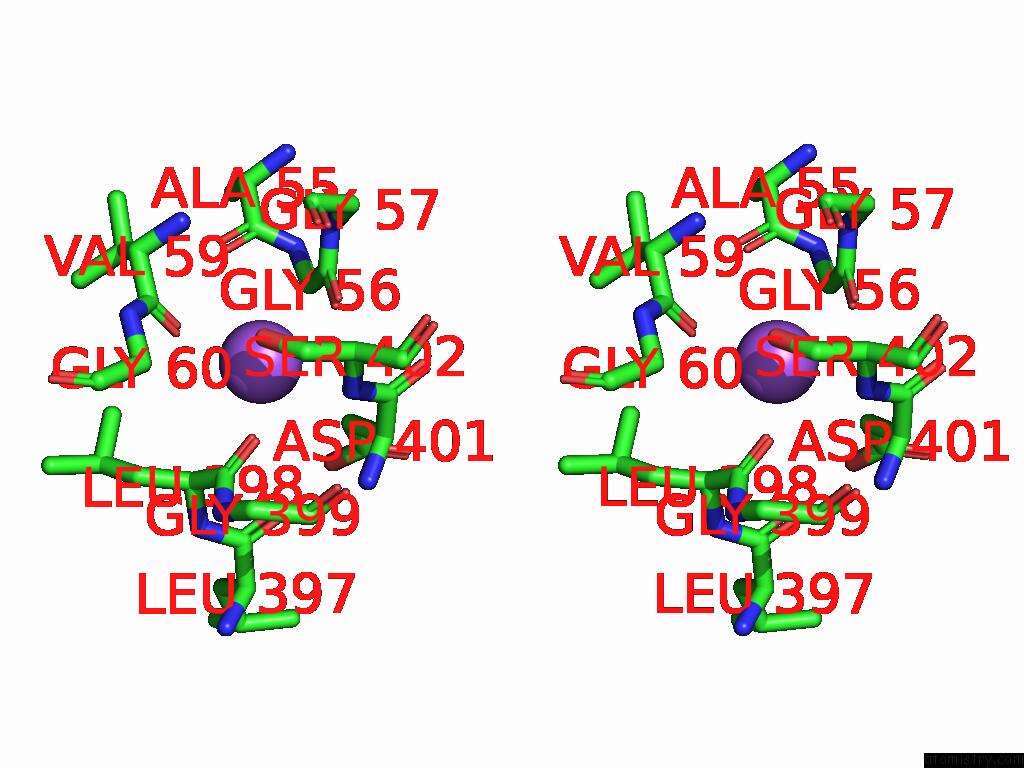
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Ges Bound Mtaut within 5.0Å range:
|
Sodium binding site 2 out of 2 in 9kyo
Go back to
Sodium binding site 2 out
of 2 in the Ges Bound Mtaut
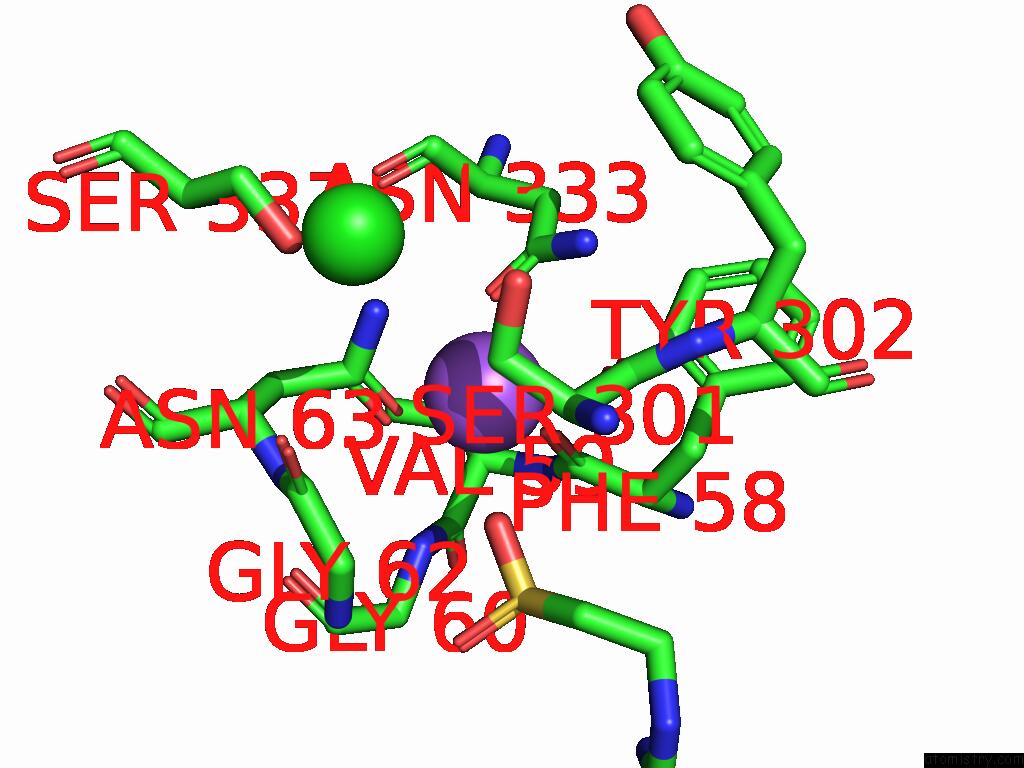
Mono view
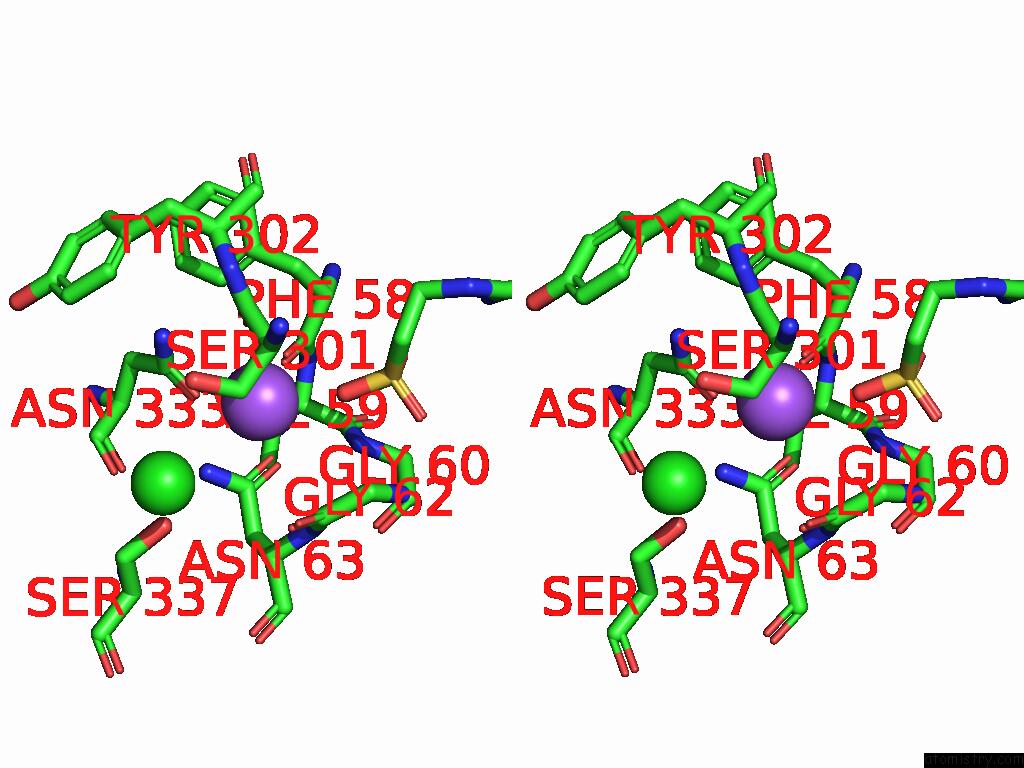
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Ges Bound Mtaut within 5.0Å range:
|
Reference:
J.She,
M.X.Wang,
J.He.
Molecular Basis For Substrate Recognition and Transport of Mammalian Taurine Transporters To Be Published.
Page generated: Mon Aug 18 17:20:45 2025
Last articles
Zn in 9QM9Zn in 9S44
Zn in 9OFE
Zn in 9OFC
Zn in 9OFD
Zn in 9OF1
Zn in 9OFB
Zn in 9N0J
Zn in 9M5X
Zn in 9LGI