Sodium »
PDB 8vnr-8wch »
8w4z »
Sodium in PDB 8w4z: Neutron Structure of Cellulase CEL6A From Phanerochaete Chrysosporium at Room Temperature, Enzyme-Product Complex, H2O Solvent
Protein crystallography data
The structure of Neutron Structure of Cellulase CEL6A From Phanerochaete Chrysosporium at Room Temperature, Enzyme-Product Complex, H2O Solvent, PDB code: 8w4z
was solved by
M.Tachioka,
S.Yamaguchi,
A.Nakamura,
T.Ishida,
K.Kusaka,
T.Yamada,
N.Yano,
T.Chatake,
T.Tamada,
K.Takeda,
S.Niwa,
H.Tanaka,
S.Takahashi,
K.Inaka,
N.Furubayashi,
S.Deguchi,
M.Samejima,
K.Igarashi,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | N/A / 1.80 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 54.73, 68.26, 89.77, 90, 90, 90 |
| R / Rfree (%) | 22.2 / 24.2 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Neutron Structure of Cellulase CEL6A From Phanerochaete Chrysosporium at Room Temperature, Enzyme-Product Complex, H2O Solvent
(pdb code 8w4z). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Neutron Structure of Cellulase CEL6A From Phanerochaete Chrysosporium at Room Temperature, Enzyme-Product Complex, H2O Solvent, PDB code: 8w4z:
In total only one binding site of Sodium was determined in the Neutron Structure of Cellulase CEL6A From Phanerochaete Chrysosporium at Room Temperature, Enzyme-Product Complex, H2O Solvent, PDB code: 8w4z:
Sodium binding site 1 out of 1 in 8w4z
Go back to
Sodium binding site 1 out
of 1 in the Neutron Structure of Cellulase CEL6A From Phanerochaete Chrysosporium at Room Temperature, Enzyme-Product Complex, H2O Solvent
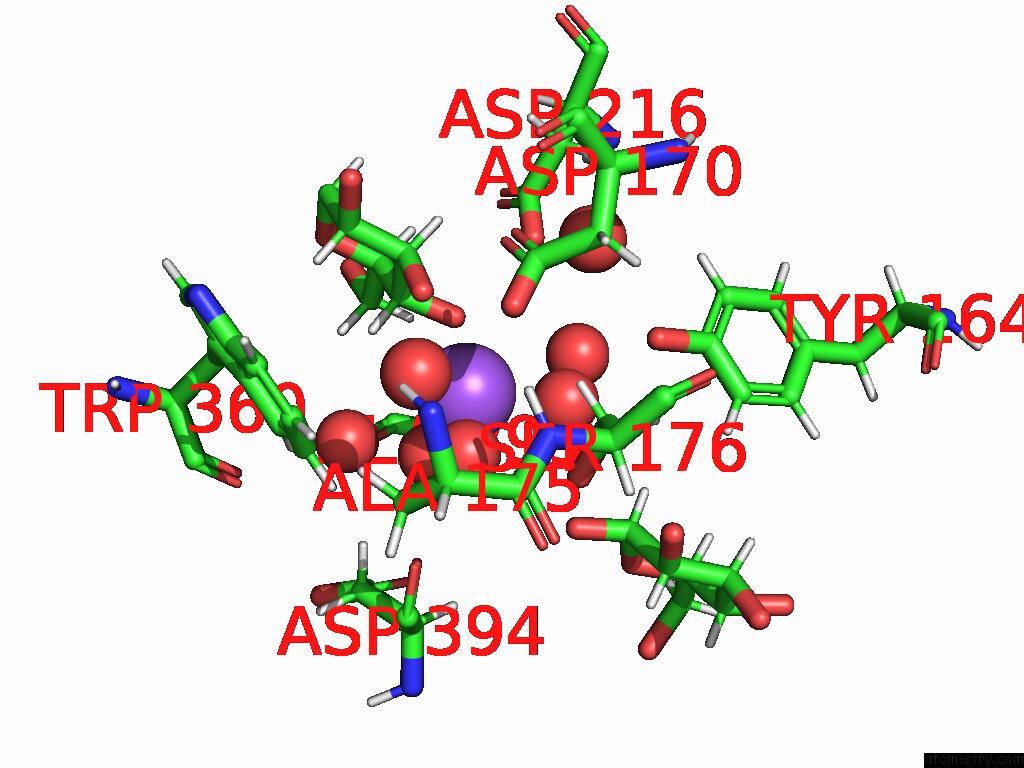
Mono view
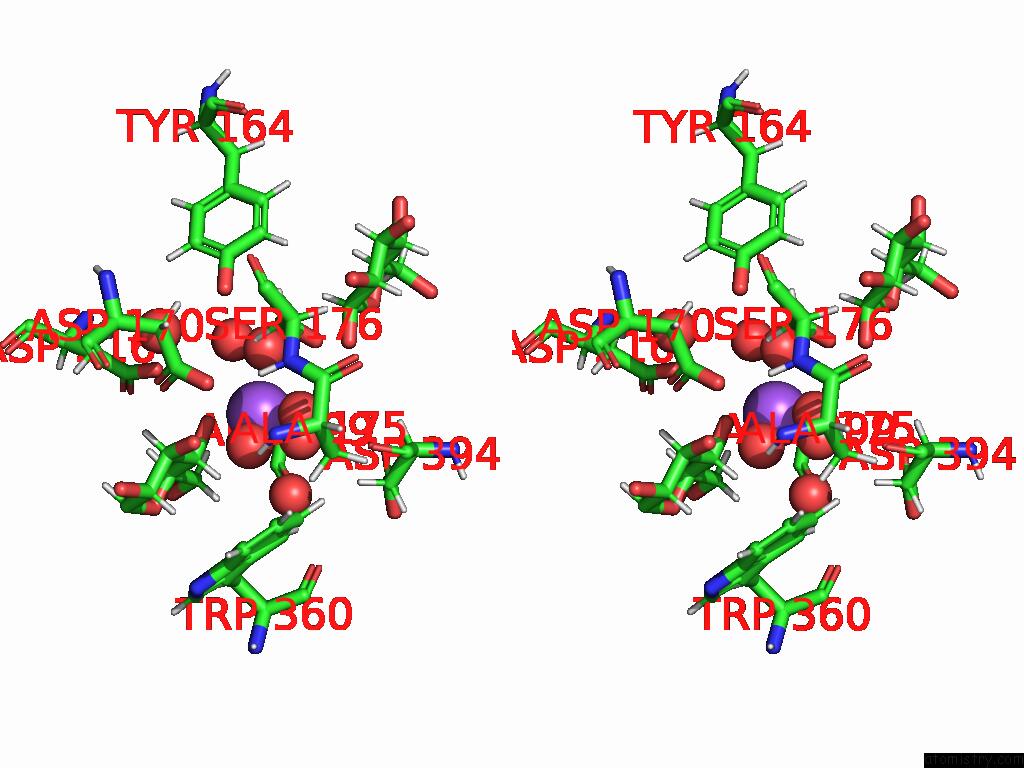
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Neutron Structure of Cellulase CEL6A From Phanerochaete Chrysosporium at Room Temperature, Enzyme-Product Complex, H2O Solvent within 5.0Å range:
|
Reference:
M.Tachioka,
S.Yamaguchi,
A.Nakamura,
T.Ishida,
K.Kusaka,
T.Yamada,
N.Yano,
T.Chatake,
T.Tamada,
K.Takeda,
S.Niwa,
H.Tanaka,
S.Takahashi,
K.Inaka,
N.Furubayashi,
S.Deguchi,
M.Samejima,
K.Igarashi.
Deprotonated Arginine Controls A Putative Catalytic Base in Invert-Ing Family 6 Glycoside Hydrolase To Be Published.
Page generated: Mon Aug 18 16:15:33 2025
Last articles
Ni in 4JPENi in 4JPC
Ni in 4JH4
Ni in 4JOO
Ni in 4JP9
Ni in 4JC7
Ni in 4J72
Ni in 4J8L
Ni in 4JCZ
Ni in 4J7Y