Sodium »
PDB 7apy-7b7n »
7avq »
Sodium in PDB 7avq: Crystal Structure of Haspin in Complex with Disubstituted Imidazo[1,2- B]Pyridazine Inhibitor (Compound 12)
Enzymatic activity of Crystal Structure of Haspin in Complex with Disubstituted Imidazo[1,2- B]Pyridazine Inhibitor (Compound 12)
All present enzymatic activity of Crystal Structure of Haspin in Complex with Disubstituted Imidazo[1,2- B]Pyridazine Inhibitor (Compound 12):
2.7.11.1;
2.7.11.1;
Protein crystallography data
The structure of Crystal Structure of Haspin in Complex with Disubstituted Imidazo[1,2- B]Pyridazine Inhibitor (Compound 12), PDB code: 7avq
was solved by
A.Chaikuad,
S.Routier,
S.Knapp,
Structural Genomics Consortium (Sgc),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 57.83 / 1.65 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 70.349, 78.009, 86.150, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 15.9 / 17.9 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Crystal Structure of Haspin in Complex with Disubstituted Imidazo[1,2- B]Pyridazine Inhibitor (Compound 12)
(pdb code 7avq). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Crystal Structure of Haspin in Complex with Disubstituted Imidazo[1,2- B]Pyridazine Inhibitor (Compound 12), PDB code: 7avq:
In total only one binding site of Sodium was determined in the Crystal Structure of Haspin in Complex with Disubstituted Imidazo[1,2- B]Pyridazine Inhibitor (Compound 12), PDB code: 7avq:
Sodium binding site 1 out of 1 in 7avq
Go back to
Sodium binding site 1 out
of 1 in the Crystal Structure of Haspin in Complex with Disubstituted Imidazo[1,2- B]Pyridazine Inhibitor (Compound 12)
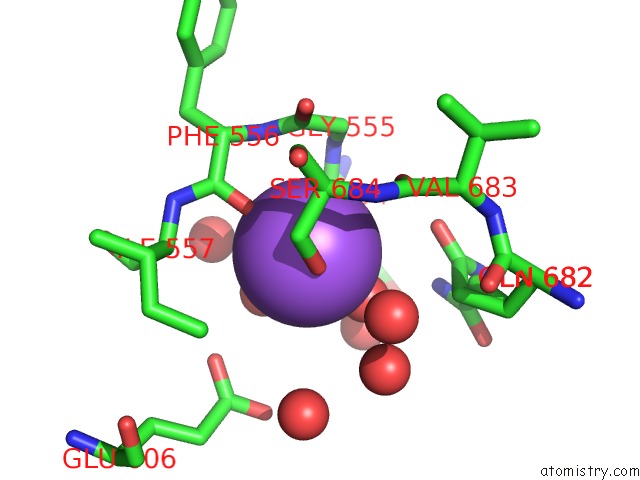
Mono view
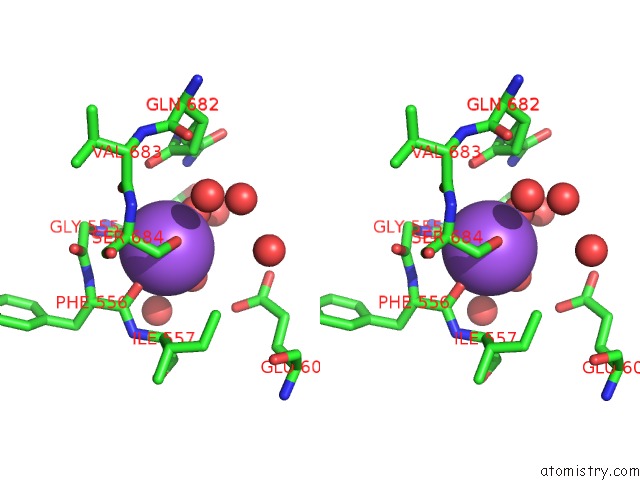
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Crystal Structure of Haspin in Complex with Disubstituted Imidazo[1,2- B]Pyridazine Inhibitor (Compound 12) within 5.0Å range:
|
Reference:
J.Elie,
O.Feizbakhsh,
N.Desban,
B.Josselin,
B.Baratte,
A.Bescond,
J.Duez,
X.Fant,
S.Bach,
D.Marie,
M.Place,
S.Ben Salah,
A.Chartier,
S.Berteina-Raboin,
A.Chaikuad,
S.Knapp,
F.Carles,
P.Bonnet,
F.Buron,
S.Routier,
S.Ruchaud.
Design of New Disubstituted Imidazo[1,2- B ]Pyridazine Derivatives As Selective Haspin Inhibitors. Synthesis, Binding Mode and Anticancer Biological Evaluation. J Enzyme Inhib Med Chem V. 35 1840 2020.
ISSN: ESSN 1475-6374
PubMed: 33040634
DOI: 10.1080/14756366.2020.1825408
Page generated: Mon Aug 18 09:12:17 2025
ISSN: ESSN 1475-6374
PubMed: 33040634
DOI: 10.1080/14756366.2020.1825408
Last articles
Na in 7UVSNa in 7USI
Na in 7UUZ
Na in 7UJJ
Na in 7USG
Na in 7URP
Na in 7UN7
Na in 7UQA
Na in 7UM2
Na in 7ULD