Sodium »
PDB 6zl4-7a2n »
6zxx »
Sodium in PDB 6zxx: Catabolic Reductive Dehalogenase Nprdha, N-Terminally Tagged.
Protein crystallography data
The structure of Catabolic Reductive Dehalogenase Nprdha, N-Terminally Tagged., PDB code: 6zxx
was solved by
D.Leys,
T.Halliwell,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 106.58 / 1.99 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 180.182, 170.391, 107.957, 90.00, 99.16, 90.00 |
| R / Rfree (%) | 18.2 / 21.8 |
Other elements in 6zxx:
The structure of Catabolic Reductive Dehalogenase Nprdha, N-Terminally Tagged. also contains other interesting chemical elements:
| Cobalt | (Co) | 3 atoms |
| Bromine | (Br) | 12 atoms |
| Iron | (Fe) | 24 atoms |
Sodium Binding Sites:
The binding sites of Sodium atom in the Catabolic Reductive Dehalogenase Nprdha, N-Terminally Tagged.
(pdb code 6zxx). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 3 binding sites of Sodium where determined in the Catabolic Reductive Dehalogenase Nprdha, N-Terminally Tagged., PDB code: 6zxx:
Jump to Sodium binding site number: 1; 2; 3;
In total 3 binding sites of Sodium where determined in the Catabolic Reductive Dehalogenase Nprdha, N-Terminally Tagged., PDB code: 6zxx:
Jump to Sodium binding site number: 1; 2; 3;
Sodium binding site 1 out of 3 in 6zxx
Go back to
Sodium binding site 1 out
of 3 in the Catabolic Reductive Dehalogenase Nprdha, N-Terminally Tagged.
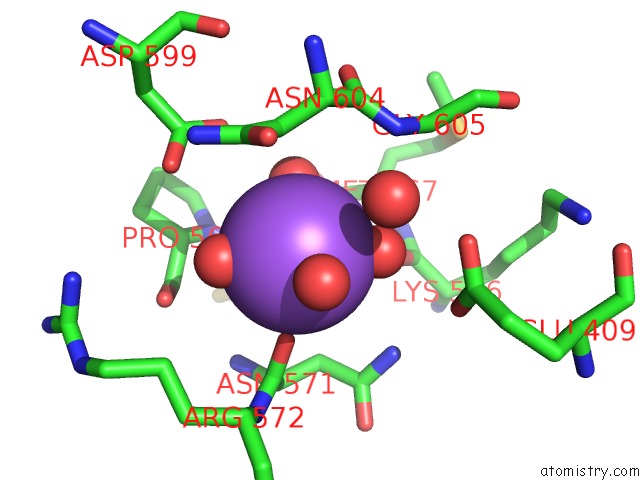
Mono view
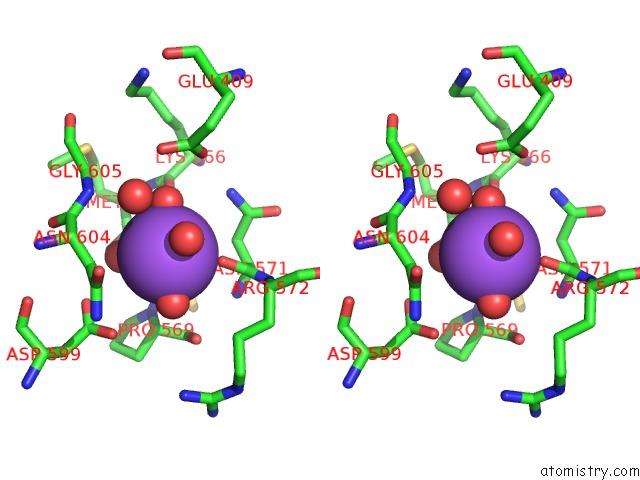
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Catabolic Reductive Dehalogenase Nprdha, N-Terminally Tagged. within 5.0Å range:
|
Sodium binding site 2 out of 3 in 6zxx
Go back to
Sodium binding site 2 out
of 3 in the Catabolic Reductive Dehalogenase Nprdha, N-Terminally Tagged.
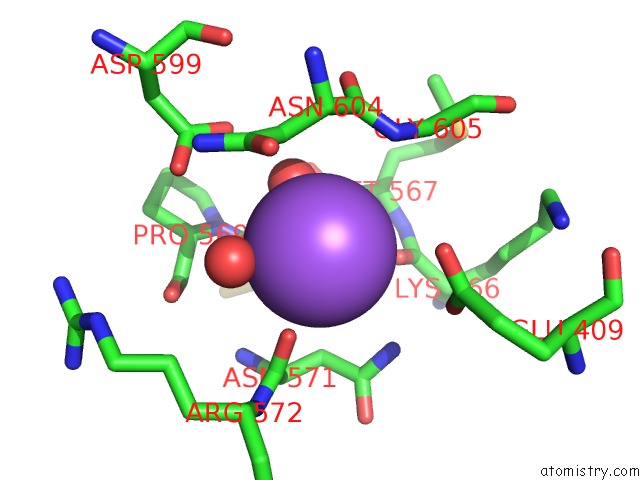
Mono view
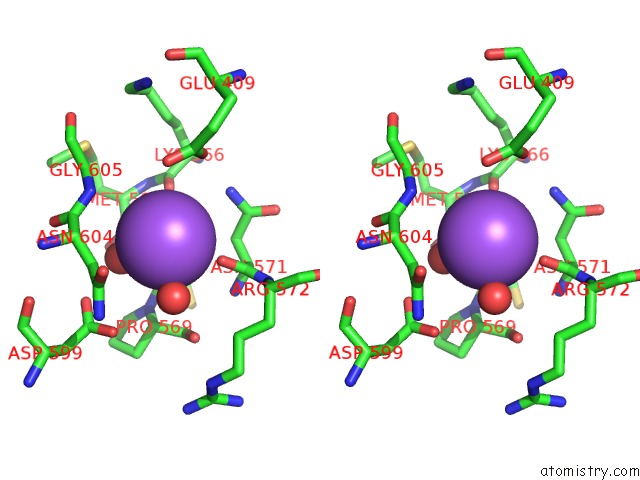
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Catabolic Reductive Dehalogenase Nprdha, N-Terminally Tagged. within 5.0Å range:
|
Sodium binding site 3 out of 3 in 6zxx
Go back to
Sodium binding site 3 out
of 3 in the Catabolic Reductive Dehalogenase Nprdha, N-Terminally Tagged.
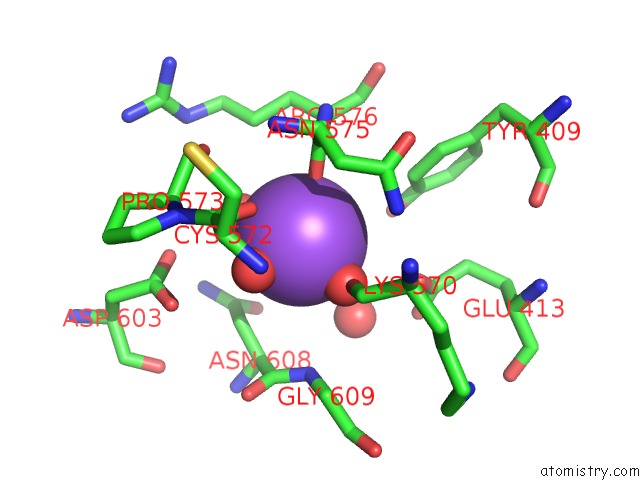
Mono view
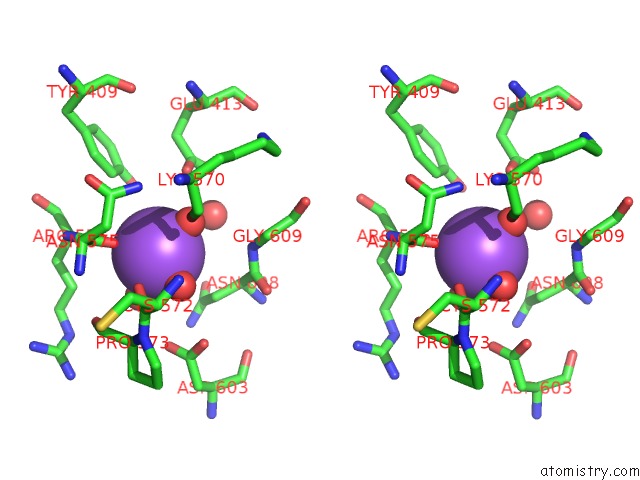
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 3 of Catabolic Reductive Dehalogenase Nprdha, N-Terminally Tagged. within 5.0Å range:
|
Reference:
T.Halliwell,
K.Fisher,
K.A.P.Payne,
S.E.J.Rigby,
D.Leys.
Catabolic Reductive Dehalogenase Substrate Complex Structures Underpin Rational Repurposing of Substrate Scope. Microorganisms V. 8 2020.
ISSN: ISSN 2076-2607
PubMed: 32887524
DOI: 10.3390/MICROORGANISMS8091344
Page generated: Tue Oct 8 15:42:32 2024
ISSN: ISSN 2076-2607
PubMed: 32887524
DOI: 10.3390/MICROORGANISMS8091344
Last articles
K in 3ZNSK in 3ZNR
K in 3ZED
K in 3WVL
K in 3ZDP
K in 3ZN2
K in 3ZDD
K in 3ZDC
K in 3ZDA
K in 3ZDB