Sodium »
PDB 6qds-6qtk »
6qf2 »
Sodium in PDB 6qf2: X-Ray Structure of Thermolysin Crystallized on A Silicon Chip
Enzymatic activity of X-Ray Structure of Thermolysin Crystallized on A Silicon Chip
All present enzymatic activity of X-Ray Structure of Thermolysin Crystallized on A Silicon Chip:
3.4.24.27;
3.4.24.27;
Protein crystallography data
The structure of X-Ray Structure of Thermolysin Crystallized on A Silicon Chip, PDB code: 6qf2
was solved by
J.Lieske,
M.Cerv,
S.Kreida,
M.Barthelmess,
P.Fischer,
T.Pakendorf,
O.Yefanov,
V.Mariani,
T.Seine,
B.H.Ross,
E.Crosas,
O.Lorbeer,
A.Burkhardt,
T.J.Lane,
S.Guenther,
J.Bergtholdt,
S.Schoen,
S.Tornroth-Horsefield,
H.N.Chapman,
A.Meents,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 46.33 / 1.73 |
| Space group | P 61 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 92.667, 92.667, 128.311, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 12.8 / 16.1 |
Other elements in 6qf2:
The structure of X-Ray Structure of Thermolysin Crystallized on A Silicon Chip also contains other interesting chemical elements:
| Zinc | (Zn) | 2 atoms |
| Calcium | (Ca) | 4 atoms |
Sodium Binding Sites:
The binding sites of Sodium atom in the X-Ray Structure of Thermolysin Crystallized on A Silicon Chip
(pdb code 6qf2). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the X-Ray Structure of Thermolysin Crystallized on A Silicon Chip, PDB code: 6qf2:
In total only one binding site of Sodium was determined in the X-Ray Structure of Thermolysin Crystallized on A Silicon Chip, PDB code: 6qf2:
Sodium binding site 1 out of 1 in 6qf2
Go back to
Sodium binding site 1 out
of 1 in the X-Ray Structure of Thermolysin Crystallized on A Silicon Chip
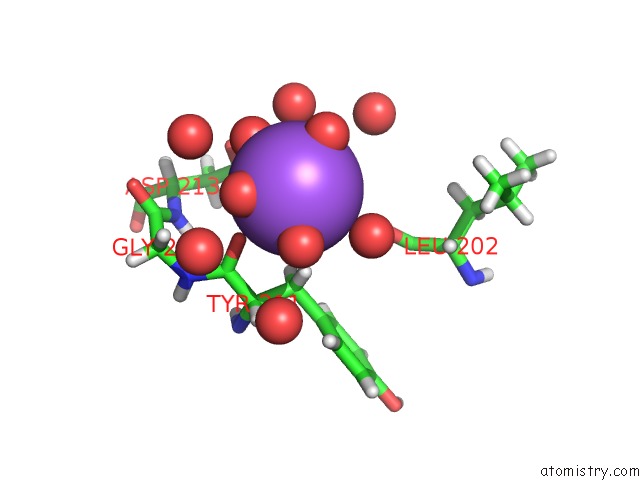
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of X-Ray Structure of Thermolysin Crystallized on A Silicon Chip within 5.0Å range:
|
Reference:
J.Lieske,
M.Cerv,
S.Kreida,
D.Komadina,
J.Fischer,
M.Barthelmess,
P.Fischer,
T.Pakendorf,
O.Yefanov,
V.Mariani,
T.Seine,
B.H.Ross,
E.Crosas,
O.Lorbeer,
A.Burkhardt,
T.J.Lane,
S.Guenther,
J.Bergtholdt,
S.Schoen,
S.Tornroth-Horsefield,
H.N.Chapman,
A.Meents.
On-Chip Crystallization For Serial Crystallography Experiments and on-Chip Ligand-Binding Studies. Iucrj V. 6 714 2019.
ISSN: ESSN 2052-2525
PubMed: 31316815
DOI: 10.1107/S2052252519007395
Page generated: Tue Oct 8 12:53:21 2024
ISSN: ESSN 2052-2525
PubMed: 31316815
DOI: 10.1107/S2052252519007395
Last articles
I in 8H5LI in 8II3
I in 8II1
I in 8IG1
I in 8GYR
I in 8GVV
I in 8GLI
I in 8GLH
I in 8EM5
I in 8GLG