Sodium »
PDB 5oqw-5pol »
5osi »
Sodium in PDB 5osi: Structure of Retromer VPS29-VPS35C Subunits Complexed with Ridl Harpin Loop (163-176)
Protein crystallography data
The structure of Structure of Retromer VPS29-VPS35C Subunits Complexed with Ridl Harpin Loop (163-176), PDB code: 5osi
was solved by
M.Romano-Moreno,
A.L.Rojas,
M.Lucas,
M.N.Isupov,
A.Hierro,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 126.49 / 2.52 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 58.554, 138.993, 126.637, 90.00, 92.78, 90.00 |
| R / Rfree (%) | 20.5 / 26.7 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Structure of Retromer VPS29-VPS35C Subunits Complexed with Ridl Harpin Loop (163-176)
(pdb code 5osi). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 4 binding sites of Sodium where determined in the Structure of Retromer VPS29-VPS35C Subunits Complexed with Ridl Harpin Loop (163-176), PDB code: 5osi:
Jump to Sodium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Sodium where determined in the Structure of Retromer VPS29-VPS35C Subunits Complexed with Ridl Harpin Loop (163-176), PDB code: 5osi:
Jump to Sodium binding site number: 1; 2; 3; 4;
Sodium binding site 1 out of 4 in 5osi
Go back to
Sodium binding site 1 out
of 4 in the Structure of Retromer VPS29-VPS35C Subunits Complexed with Ridl Harpin Loop (163-176)
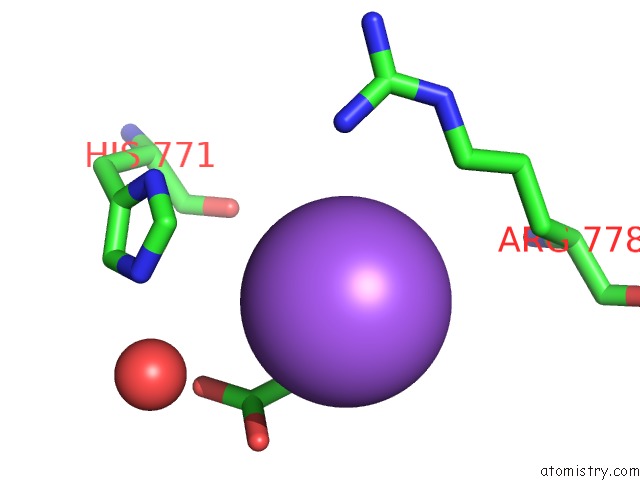
Mono view
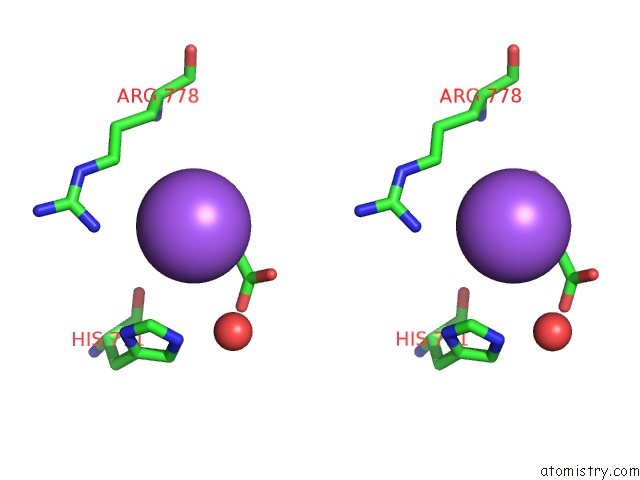
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Structure of Retromer VPS29-VPS35C Subunits Complexed with Ridl Harpin Loop (163-176) within 5.0Å range:
|
Sodium binding site 2 out of 4 in 5osi
Go back to
Sodium binding site 2 out
of 4 in the Structure of Retromer VPS29-VPS35C Subunits Complexed with Ridl Harpin Loop (163-176)
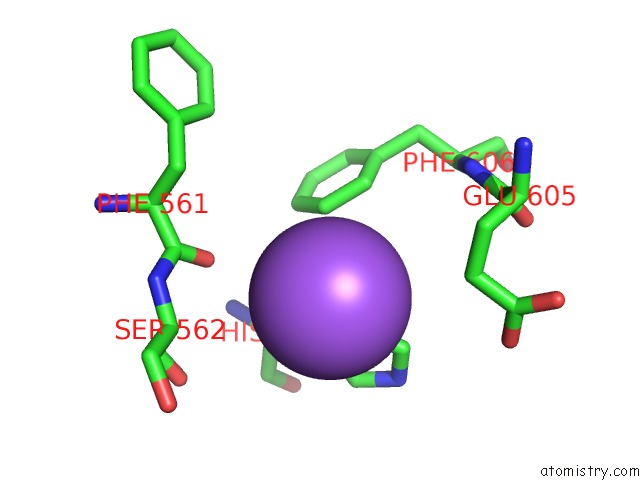
Mono view
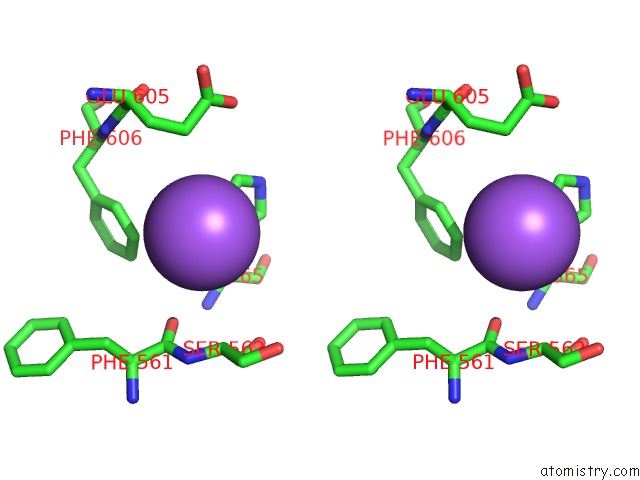
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Structure of Retromer VPS29-VPS35C Subunits Complexed with Ridl Harpin Loop (163-176) within 5.0Å range:
|
Sodium binding site 3 out of 4 in 5osi
Go back to
Sodium binding site 3 out
of 4 in the Structure of Retromer VPS29-VPS35C Subunits Complexed with Ridl Harpin Loop (163-176)
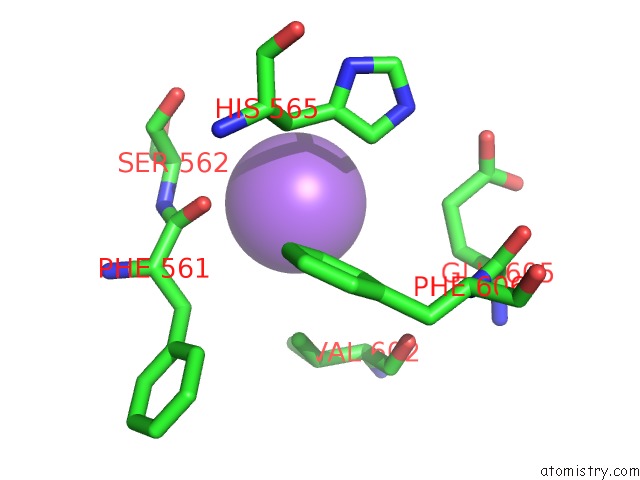
Mono view
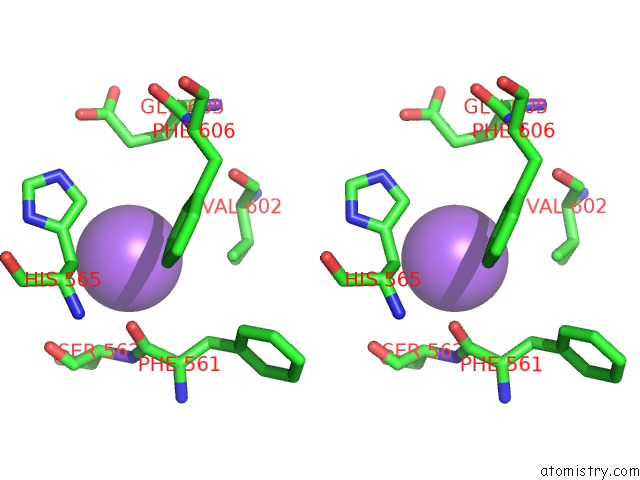
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 3 of Structure of Retromer VPS29-VPS35C Subunits Complexed with Ridl Harpin Loop (163-176) within 5.0Å range:
|
Sodium binding site 4 out of 4 in 5osi
Go back to
Sodium binding site 4 out
of 4 in the Structure of Retromer VPS29-VPS35C Subunits Complexed with Ridl Harpin Loop (163-176)
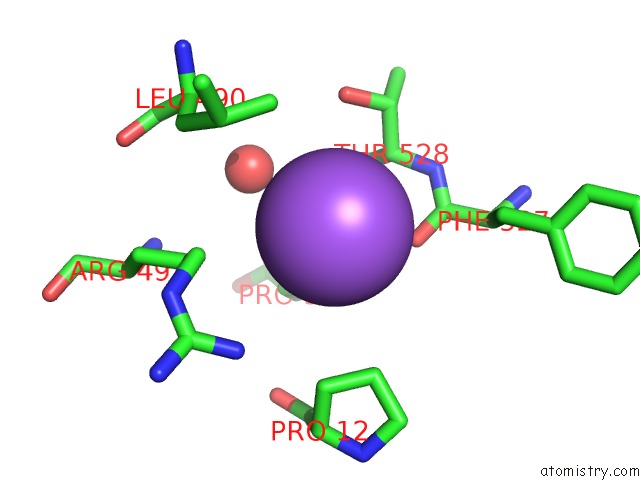
Mono view
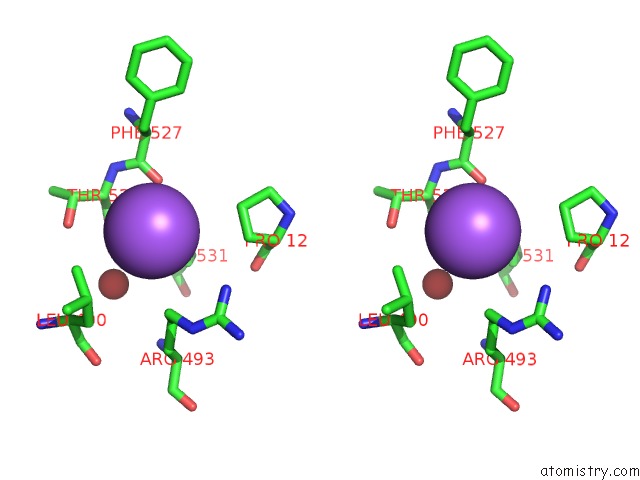
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 4 of Structure of Retromer VPS29-VPS35C Subunits Complexed with Ridl Harpin Loop (163-176) within 5.0Å range:
|
Reference:
M.Romano-Moreno,
A.L.Rojas,
C.D.Williamson,
D.C.Gershlick,
M.Lucas,
M.N.Isupov,
J.S.Bonifacino,
M.P.Machner,
A.Hierro.
Molecular Mechanism For the Subversion of the Retromer Coat By the Legionella Effector Ridl. Proc. Natl. Acad. Sci. V. 114 11151 2017U.S.A..
ISSN: ESSN 1091-6490
PubMed: 29229824
DOI: 10.1073/PNAS.1715361115
Page generated: Mon Oct 7 23:15:07 2024
ISSN: ESSN 1091-6490
PubMed: 29229824
DOI: 10.1073/PNAS.1715361115
Last articles
K in 8K1QK in 8K1J
K in 8K1E
K in 8JZG
K in 8K0U
K in 8K0T
K in 8JGW
K in 8JZA
K in 8JC7
K in 8JGR