Sodium »
PDB 8wci-8xwo »
8wt1 »
Sodium in PDB 8wt1: Crystal Structure of S9 Carboxypeptidase From Geobacillus Sterothermophilus
Protein crystallography data
The structure of Crystal Structure of S9 Carboxypeptidase From Geobacillus Sterothermophilus, PDB code: 8wt1
was solved by
K.Chandravanshi,
A.Kumar,
C.Sen,
R.Singh,
G.B.Bhange,
R.D.Makde,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.94 / 2.00 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 194.123, 75.443, 219.74, 90, 93.27, 90 |
| R / Rfree (%) | 21.2 / 25.6 |
Sodium Binding Sites:
Pages:
>>> Page 1 <<< Page 2, Binding sites: 11 - 14;Binding sites:
The binding sites of Sodium atom in the Crystal Structure of S9 Carboxypeptidase From Geobacillus Sterothermophilus (pdb code 8wt1). This binding sites where shown within 5.0 Angstroms radius around Sodium atom.In total 14 binding sites of Sodium where determined in the Crystal Structure of S9 Carboxypeptidase From Geobacillus Sterothermophilus, PDB code: 8wt1:
Jump to Sodium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Sodium binding site 1 out of 14 in 8wt1
Go back to
Sodium binding site 1 out
of 14 in the Crystal Structure of S9 Carboxypeptidase From Geobacillus Sterothermophilus
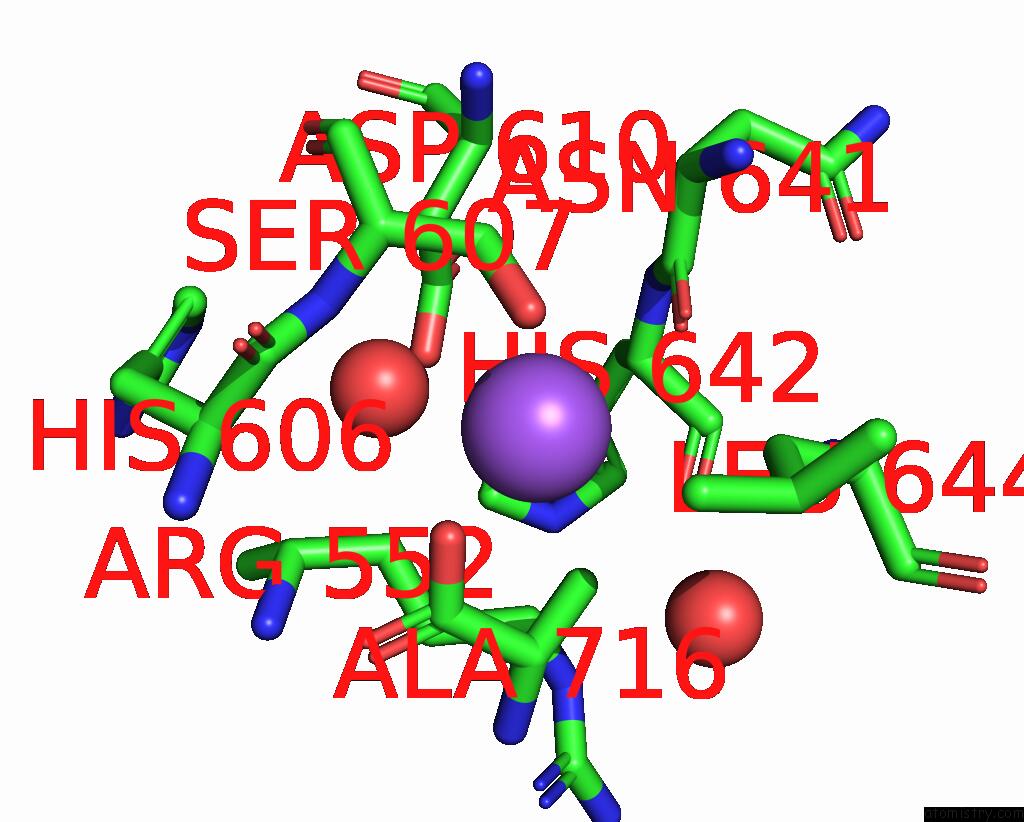
Mono view
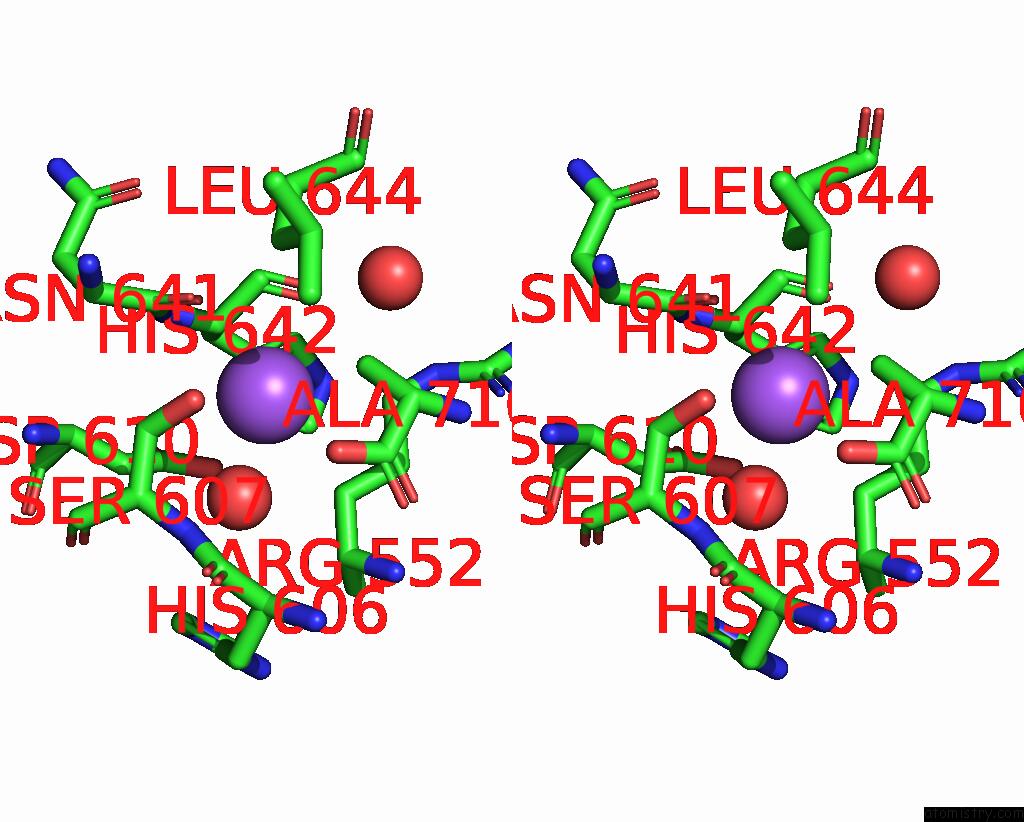
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Crystal Structure of S9 Carboxypeptidase From Geobacillus Sterothermophilus within 5.0Å range:
|
Sodium binding site 2 out of 14 in 8wt1
Go back to
Sodium binding site 2 out
of 14 in the Crystal Structure of S9 Carboxypeptidase From Geobacillus Sterothermophilus
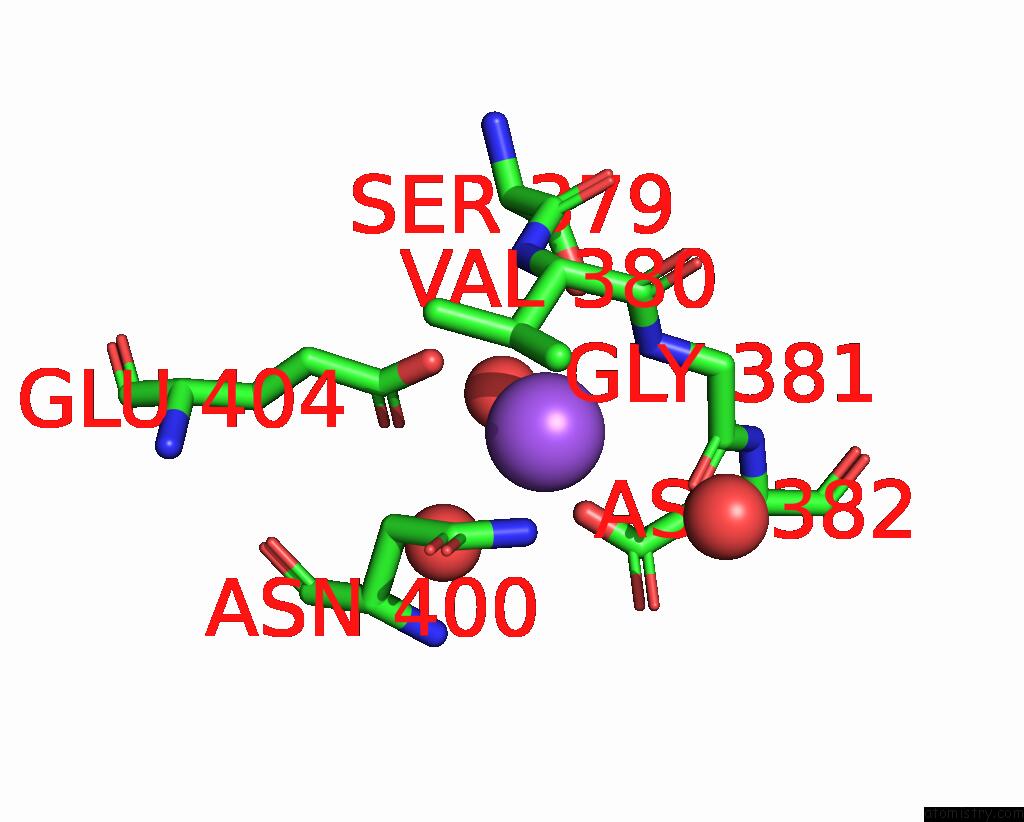
Mono view
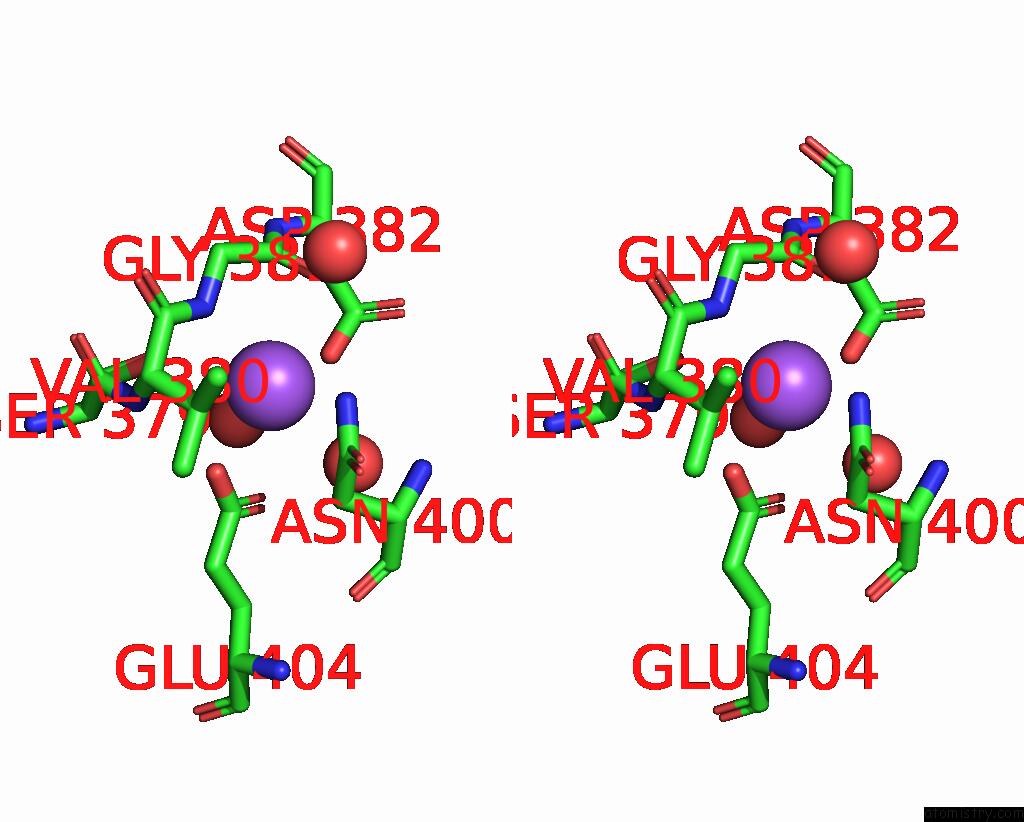
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Crystal Structure of S9 Carboxypeptidase From Geobacillus Sterothermophilus within 5.0Å range:
|
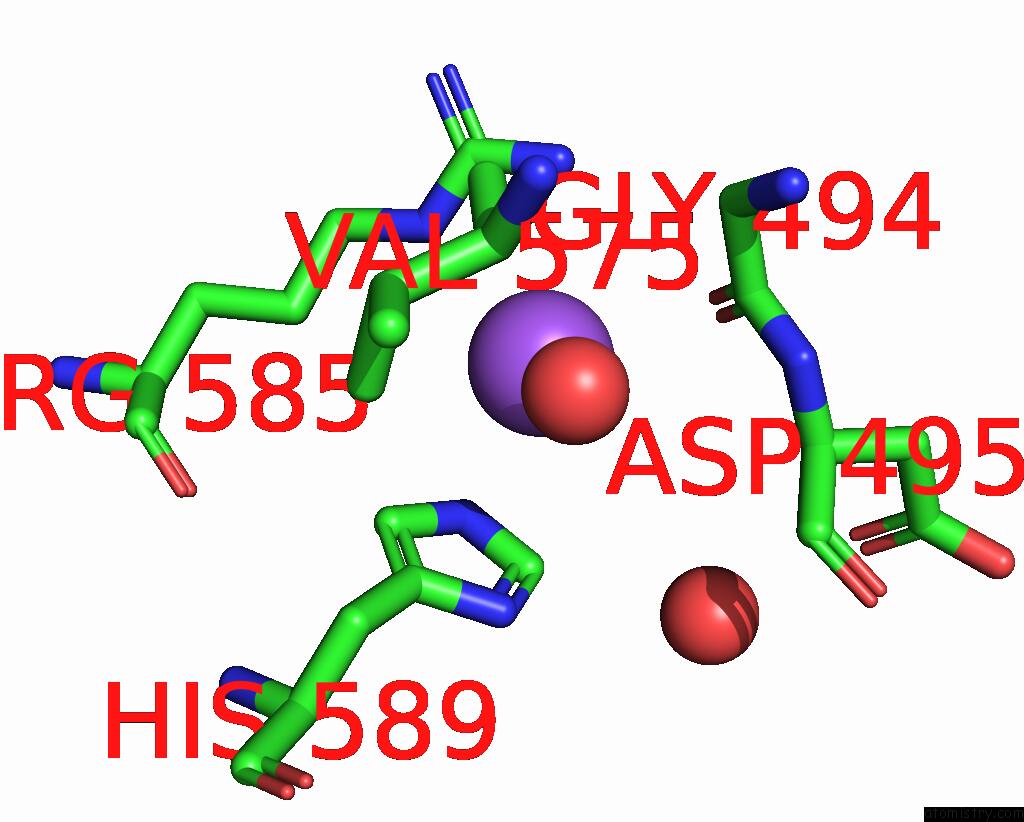
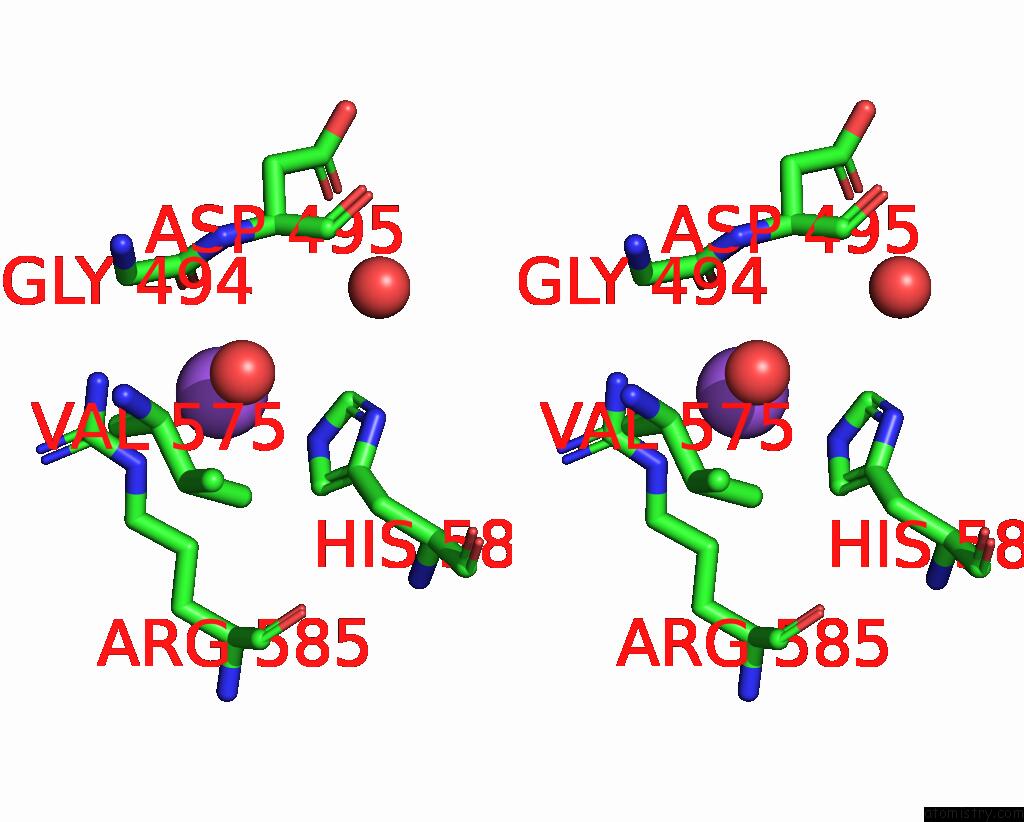
Sodium binding site 3 out of 14 in 8wt1
Go back to
Sodium binding site 3 out
of 14 in the Crystal Structure of S9 Carboxypeptidase From Geobacillus Sterothermophilus
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 3 of Crystal Structure of S9 Carboxypeptidase From Geobacillus Sterothermophilus within 5.0Å range:
|
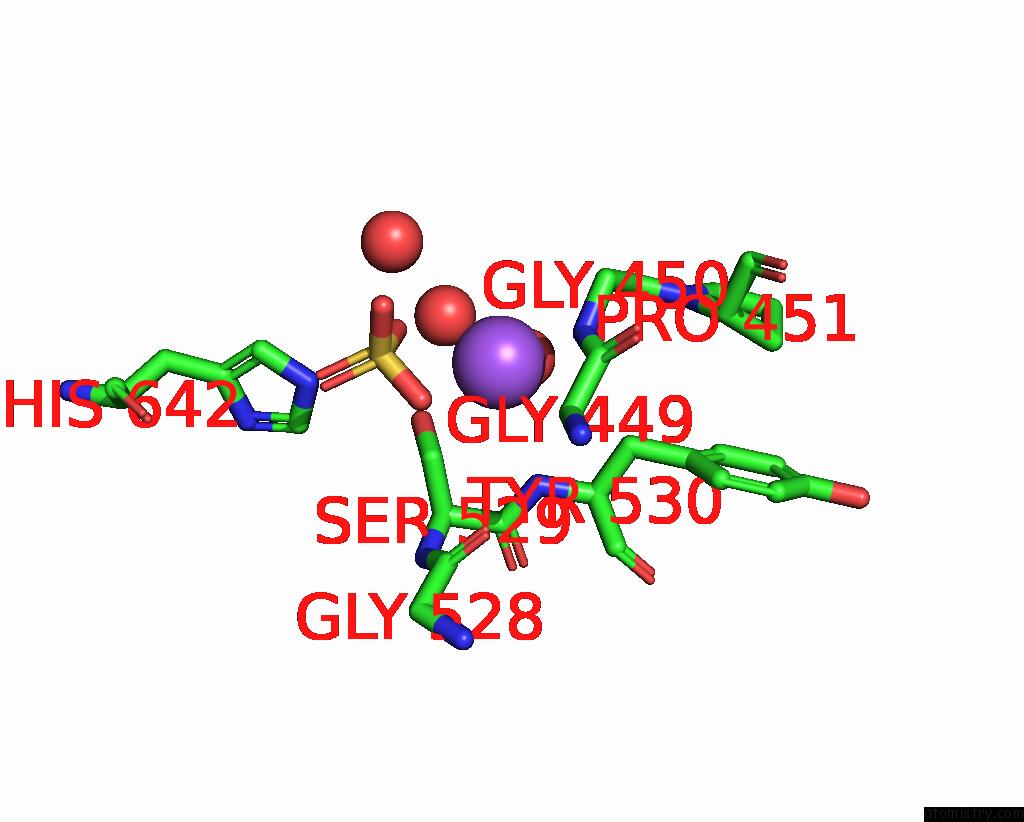
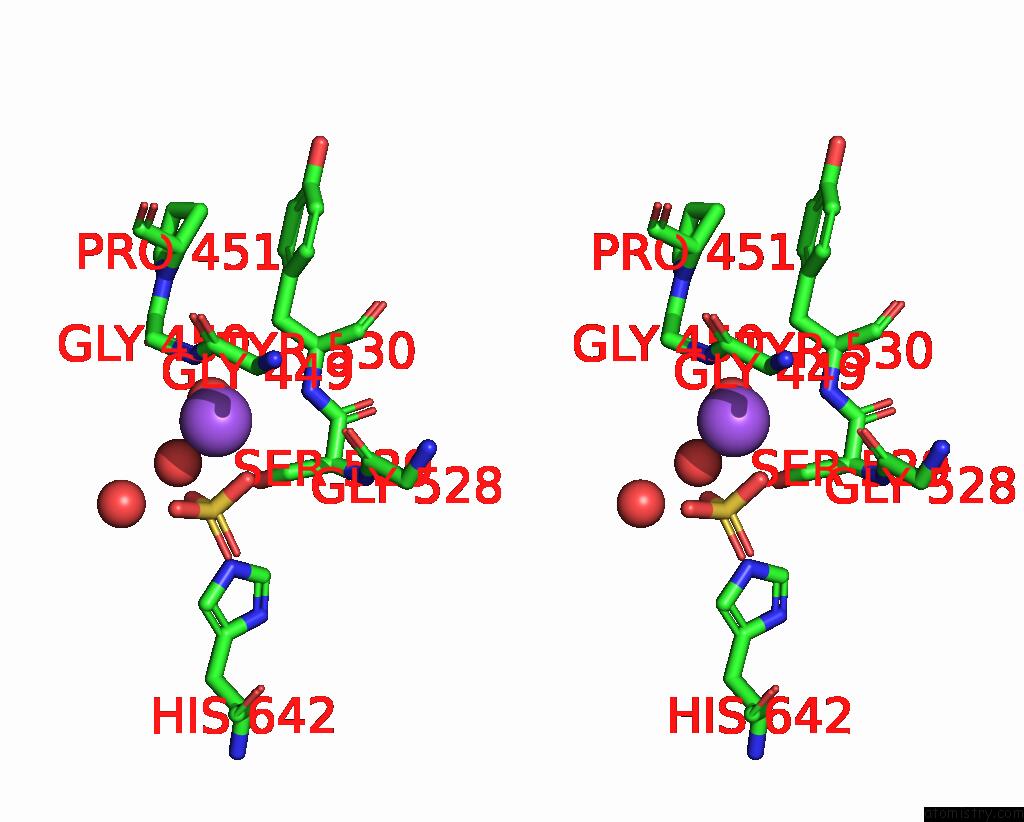
Sodium binding site 4 out of 14 in 8wt1
Go back to
Sodium binding site 4 out
of 14 in the Crystal Structure of S9 Carboxypeptidase From Geobacillus Sterothermophilus
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 4 of Crystal Structure of S9 Carboxypeptidase From Geobacillus Sterothermophilus within 5.0Å range:
|
Sodium binding site 5 out of 14 in 8wt1
Go back to
Sodium binding site 5 out
of 14 in the Crystal Structure of S9 Carboxypeptidase From Geobacillus Sterothermophilus
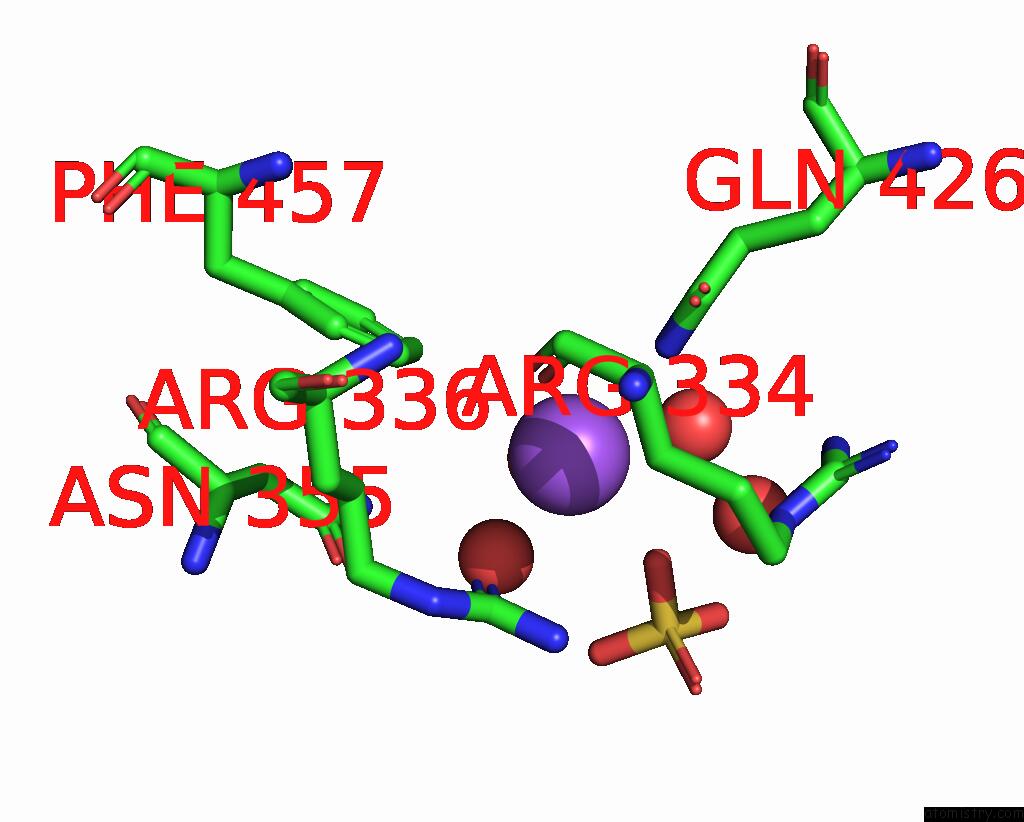
Mono view
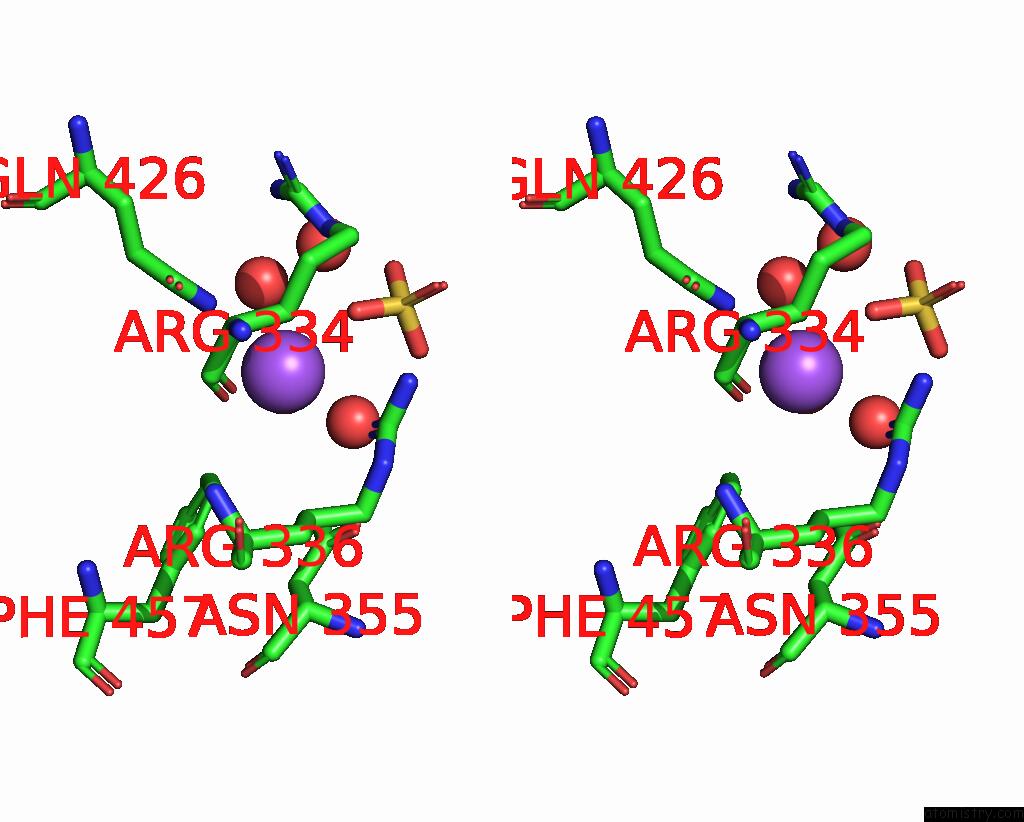
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 5 of Crystal Structure of S9 Carboxypeptidase From Geobacillus Sterothermophilus within 5.0Å range:
|
Sodium binding site 6 out of 14 in 8wt1
Go back to
Sodium binding site 6 out
of 14 in the Crystal Structure of S9 Carboxypeptidase From Geobacillus Sterothermophilus
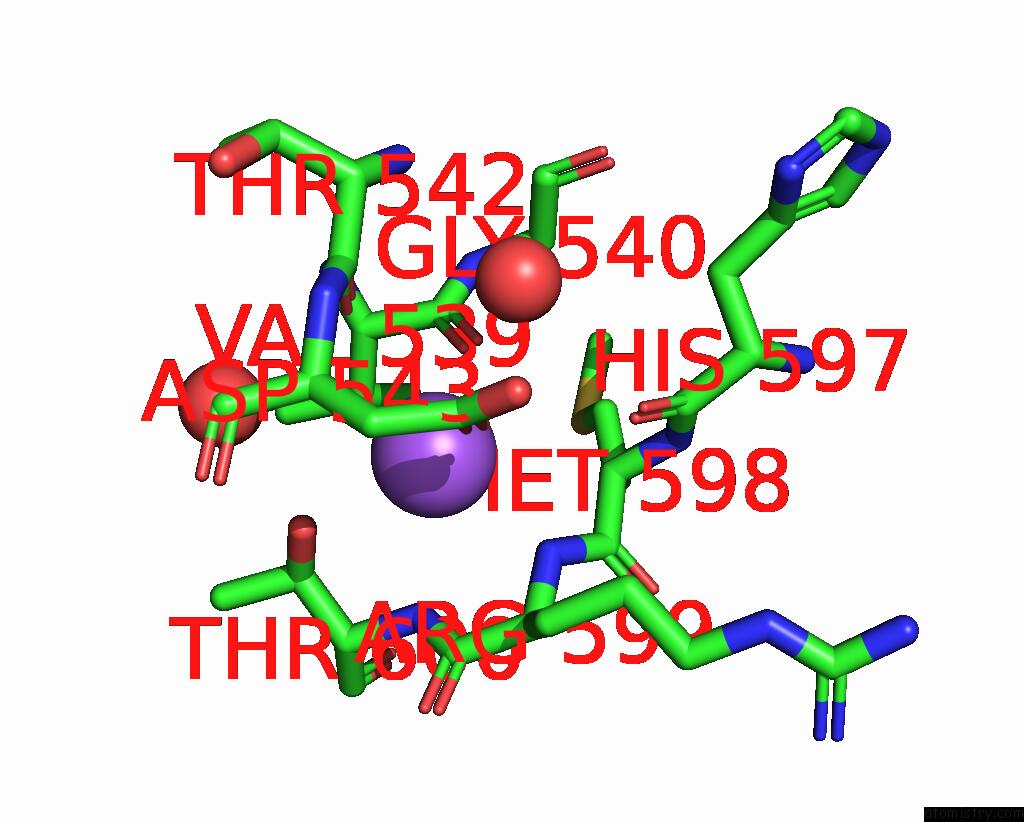
Mono view
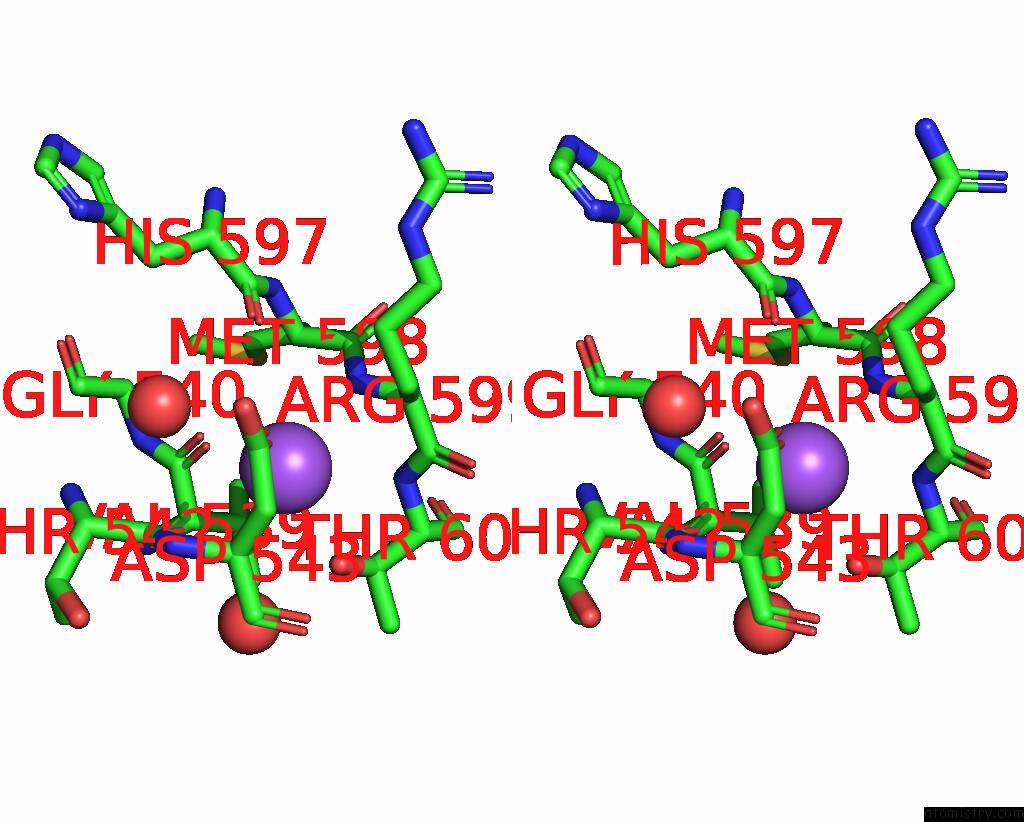
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 6 of Crystal Structure of S9 Carboxypeptidase From Geobacillus Sterothermophilus within 5.0Å range:
|
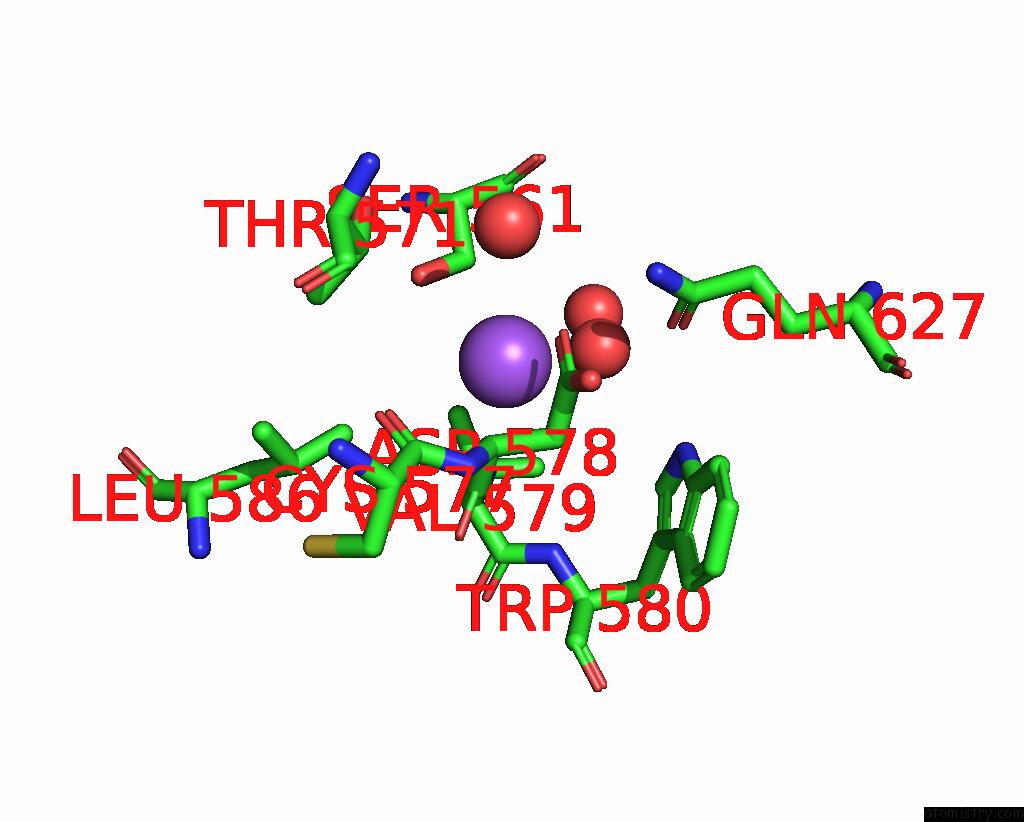
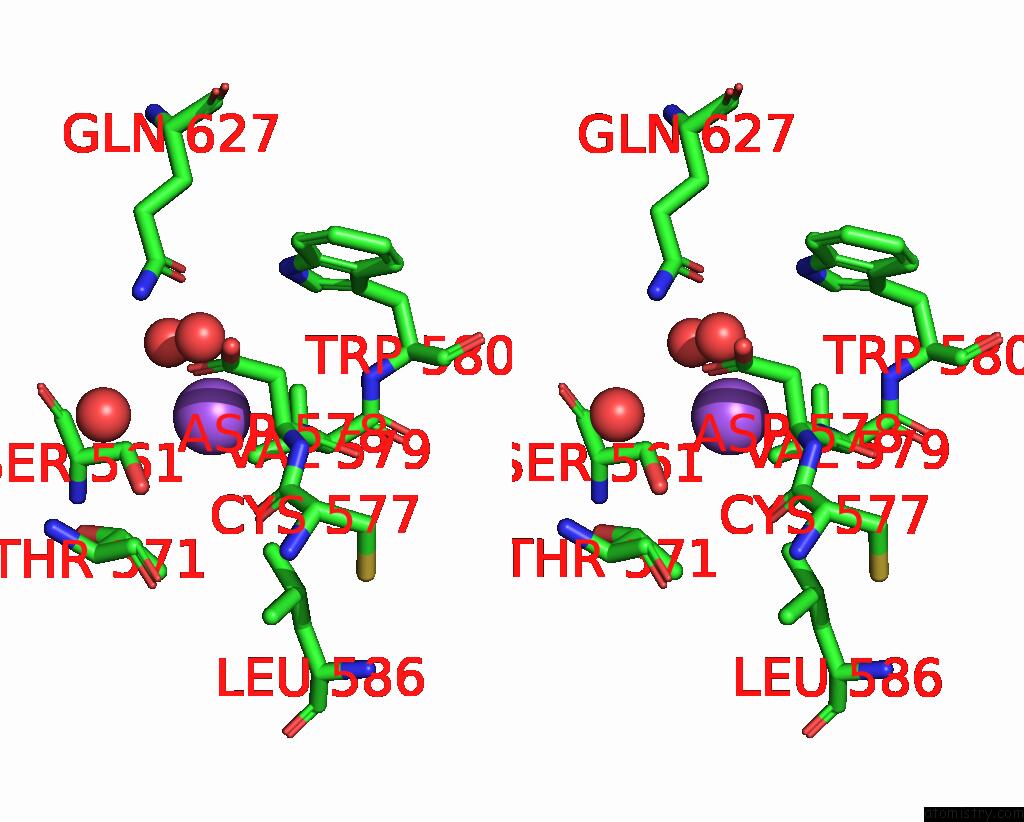
Sodium binding site 7 out of 14 in 8wt1
Go back to
Sodium binding site 7 out
of 14 in the Crystal Structure of S9 Carboxypeptidase From Geobacillus Sterothermophilus
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 7 of Crystal Structure of S9 Carboxypeptidase From Geobacillus Sterothermophilus within 5.0Å range:
|
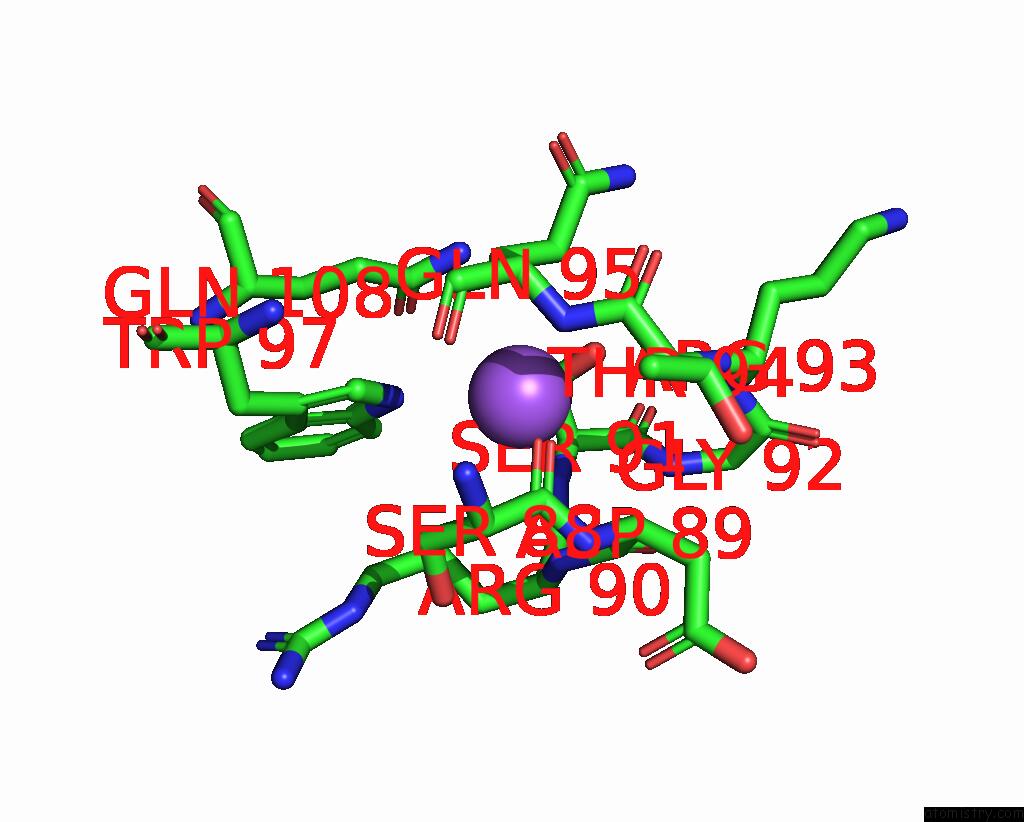
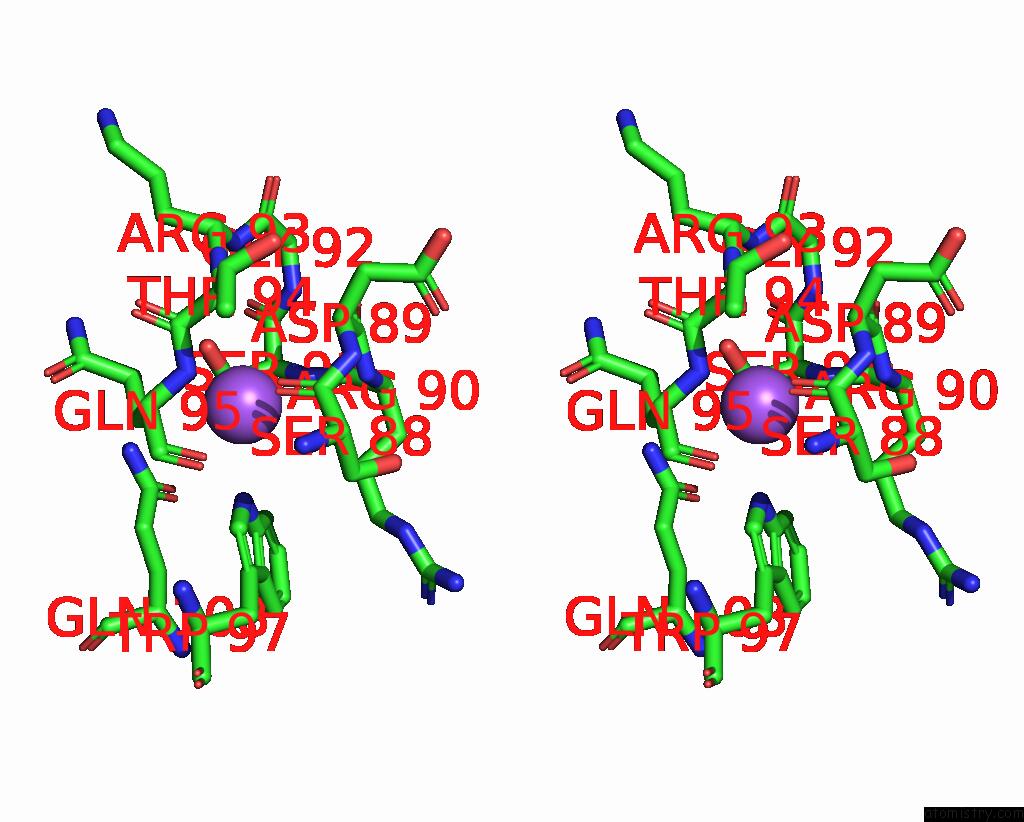
Sodium binding site 8 out of 14 in 8wt1
Go back to
Sodium binding site 8 out
of 14 in the Crystal Structure of S9 Carboxypeptidase From Geobacillus Sterothermophilus
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 8 of Crystal Structure of S9 Carboxypeptidase From Geobacillus Sterothermophilus within 5.0Å range:
|
Sodium binding site 9 out of 14 in 8wt1
Go back to
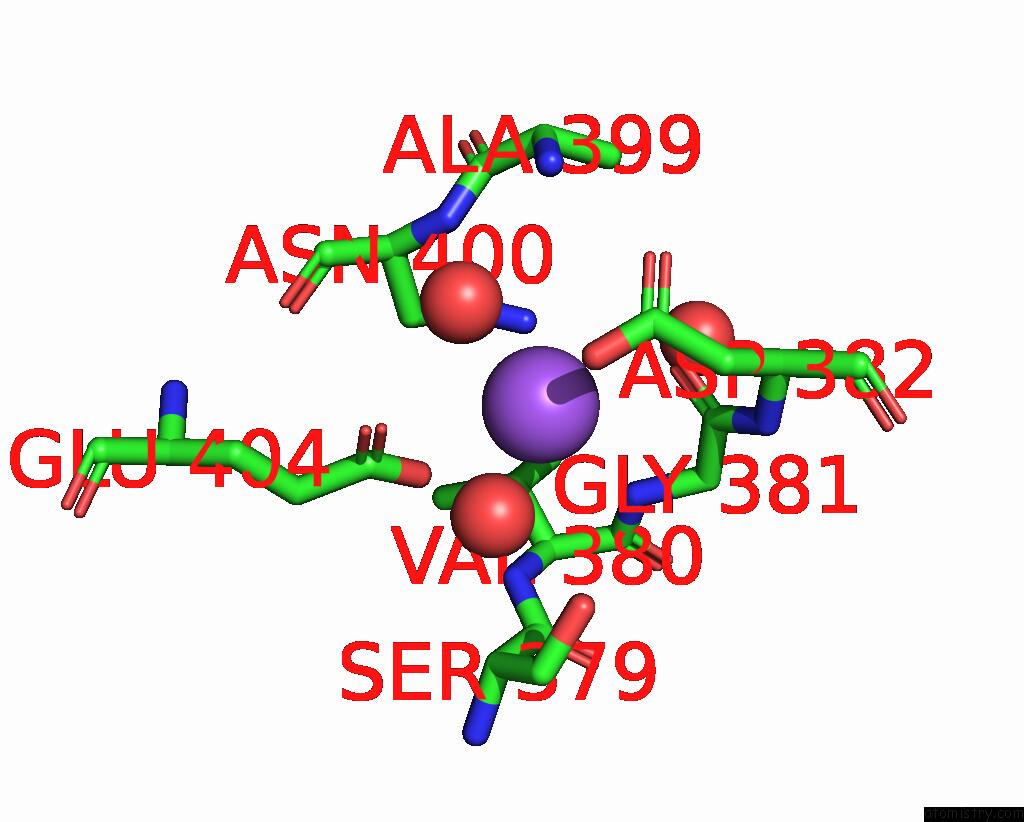
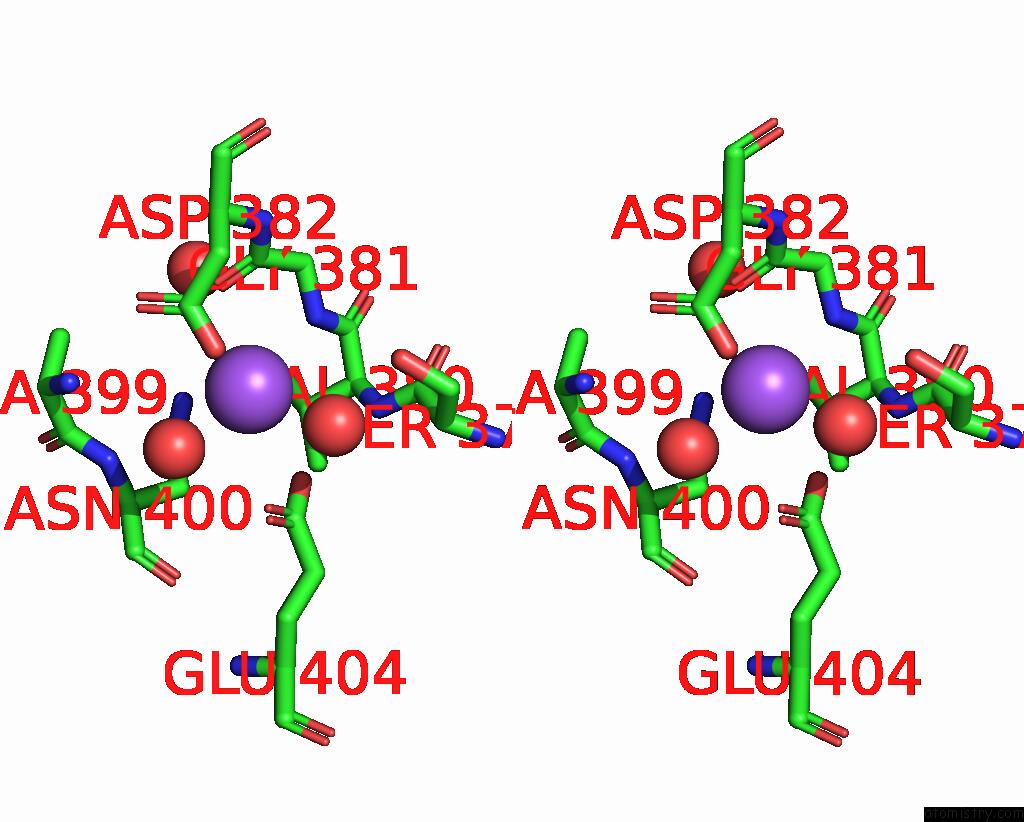
Sodium binding site 9 out
of 14 in the Crystal Structure of S9 Carboxypeptidase From Geobacillus Sterothermophilus
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 9 of Crystal Structure of S9 Carboxypeptidase From Geobacillus Sterothermophilus within 5.0Å range:
|
Sodium binding site 10 out of 14 in 8wt1
Go back to
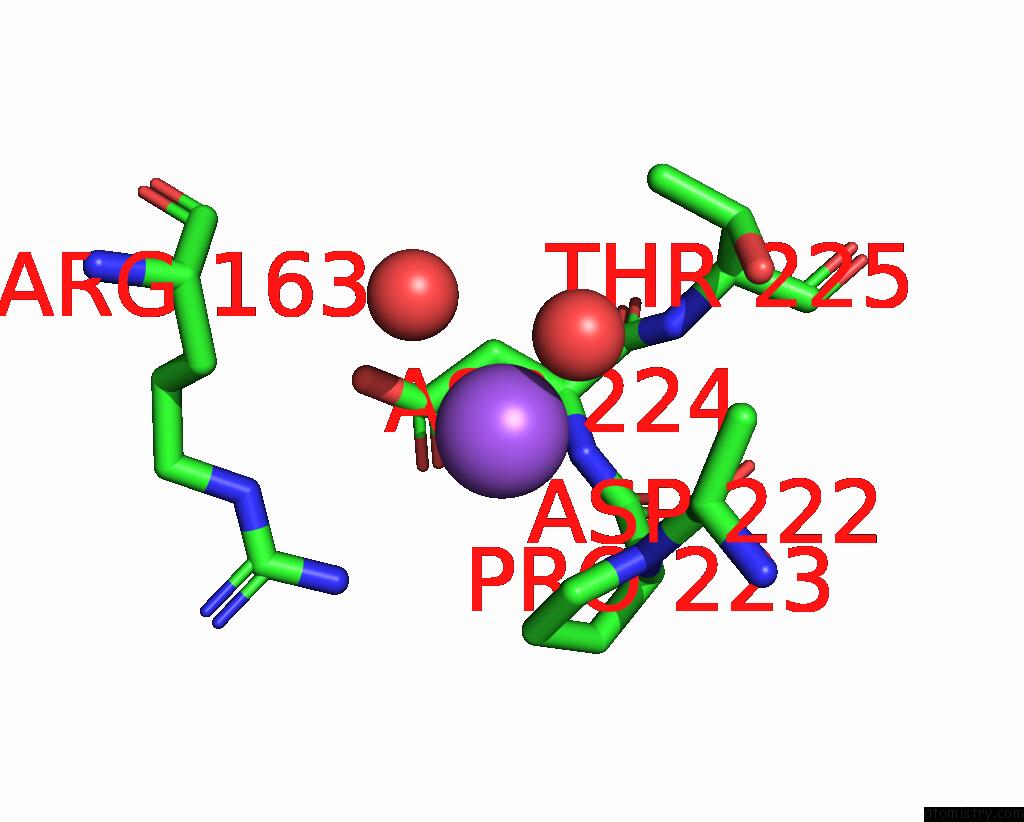
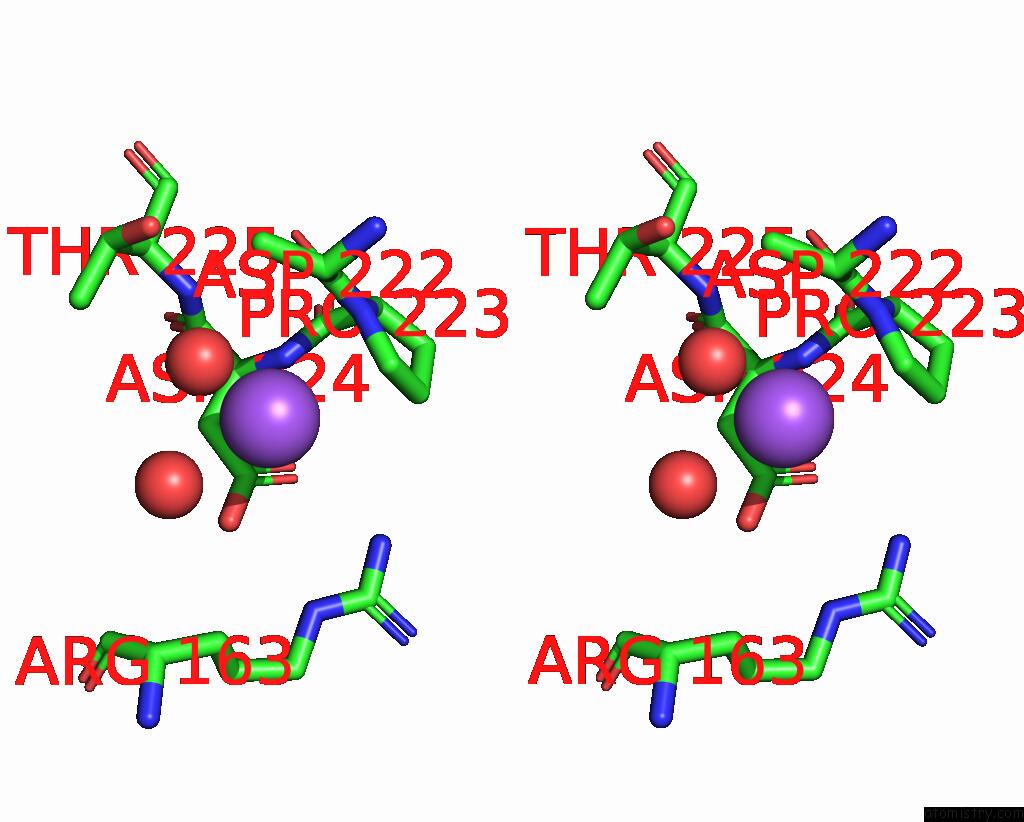
Sodium binding site 10 out
of 14 in the Crystal Structure of S9 Carboxypeptidase From Geobacillus Sterothermophilus
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 10 of Crystal Structure of S9 Carboxypeptidase From Geobacillus Sterothermophilus within 5.0Å range:
|
Reference:
K.Chandravanshi,
R.Singh,
G.N.Bhange,
A.Kumar,
P.Yadav,
A.Kumar,
R.D.Makde.
Crystal Structure and Solution Scattering of Geobacillus Stearothermophilus S9 Peptidase Reveal Structural Adaptations For Carboxypeptidase Activity. Febs Lett. 2024.
ISSN: ISSN 0014-5793
PubMed: 38426217
DOI: 10.1002/1873-3468.14834
Page generated: Mon Aug 18 16:20:23 2025
ISSN: ISSN 0014-5793
PubMed: 38426217
DOI: 10.1002/1873-3468.14834
Last articles
Ni in 7ODHNi in 7OLL
Ni in 7OLK
Ni in 7O9C
Ni in 7O9E
Ni in 7ODG
Ni in 7ODF
Ni in 7O54
Ni in 7NYS
Ni in 7O4Z