Sodium »
PDB 7ugj-7vm9 »
7v8s »
Sodium in PDB 7v8s: Crystal Structure of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed with Nadp+ and Fad in Space Group of P1211
Enzymatic activity of Crystal Structure of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed with Nadp+ and Fad in Space Group of P1211
All present enzymatic activity of Crystal Structure of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed with Nadp+ and Fad in Space Group of P1211:
1.14.13.22;
1.14.13.22;
Protein crystallography data
The structure of Crystal Structure of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed with Nadp+ and Fad in Space Group of P1211, PDB code: 7v8s
was solved by
T.Li,
G.Y.Li,
H.Yin,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 27.79 / 2.08 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 61.122, 164.034, 62.052, 90, 115.88, 90 |
| R / Rfree (%) | 17.2 / 21.8 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Crystal Structure of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed with Nadp+ and Fad in Space Group of P1211
(pdb code 7v8s). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 5 binding sites of Sodium where determined in the Crystal Structure of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed with Nadp+ and Fad in Space Group of P1211, PDB code: 7v8s:
Jump to Sodium binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Sodium where determined in the Crystal Structure of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed with Nadp+ and Fad in Space Group of P1211, PDB code: 7v8s:
Jump to Sodium binding site number: 1; 2; 3; 4; 5;
Sodium binding site 1 out of 5 in 7v8s
Go back to
Sodium binding site 1 out
of 5 in the Crystal Structure of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed with Nadp+ and Fad in Space Group of P1211
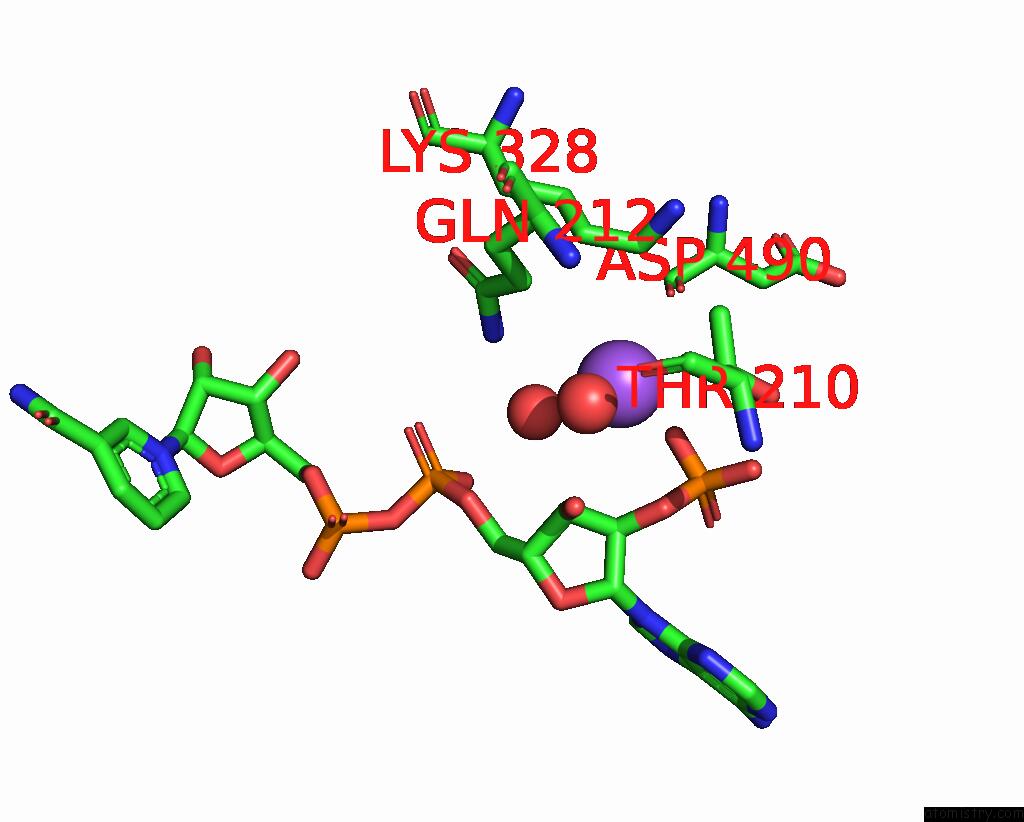
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Crystal Structure of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed with Nadp+ and Fad in Space Group of P1211 within 5.0Å range:
|
Sodium binding site 2 out of 5 in 7v8s
Go back to
Sodium binding site 2 out
of 5 in the Crystal Structure of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed with Nadp+ and Fad in Space Group of P1211
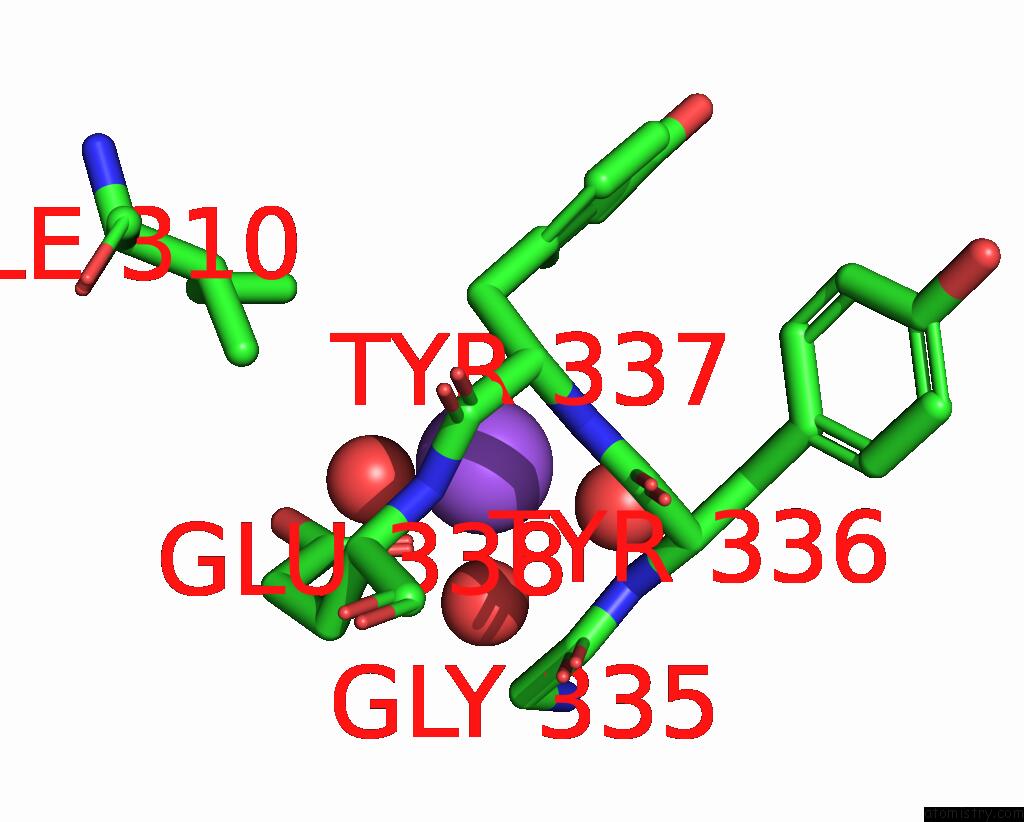
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Crystal Structure of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed with Nadp+ and Fad in Space Group of P1211 within 5.0Å range:
|
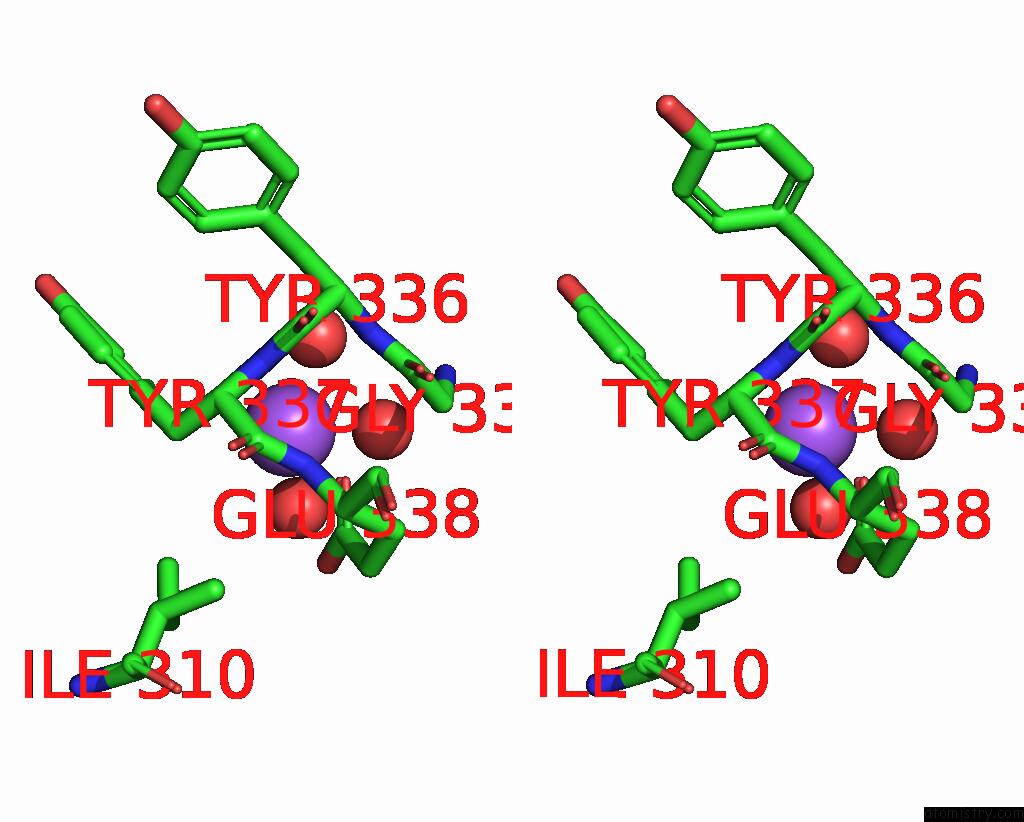
Sodium binding site 3 out of 5 in 7v8s
Go back to
Sodium binding site 3 out
of 5 in the Crystal Structure of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed with Nadp+ and Fad in Space Group of P1211

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 3 of Crystal Structure of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed with Nadp+ and Fad in Space Group of P1211 within 5.0Å range:
|
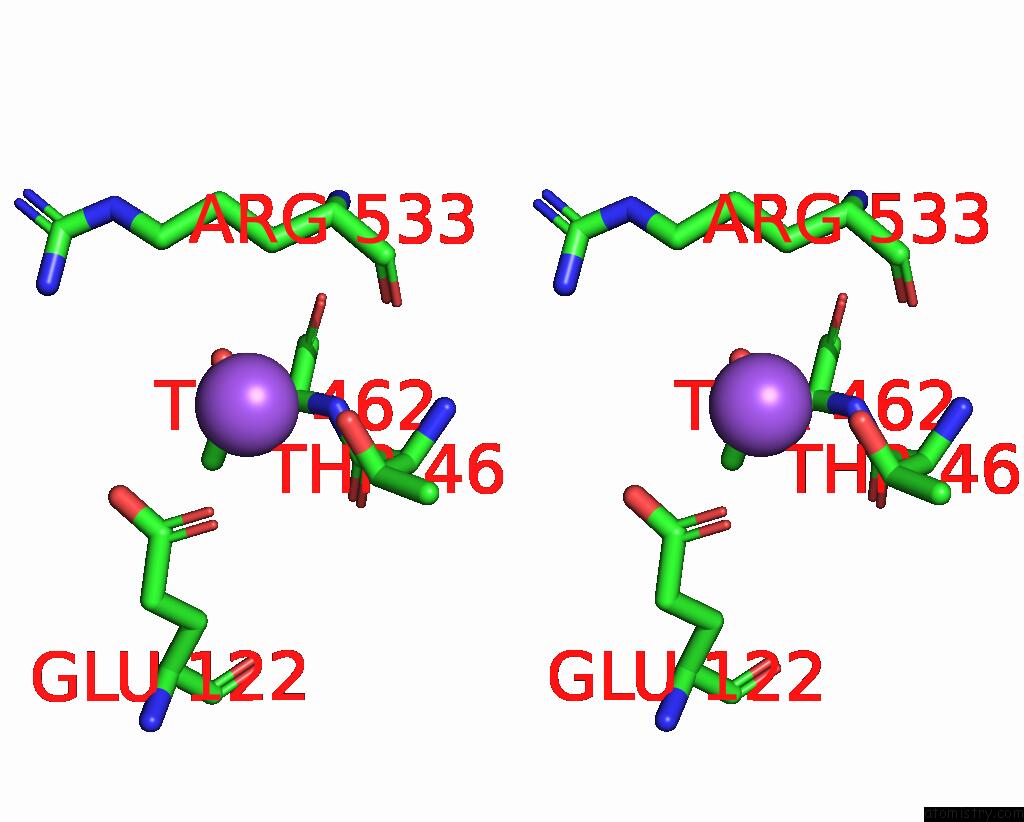
Sodium binding site 4 out of 5 in 7v8s
Go back to
Sodium binding site 4 out
of 5 in the Crystal Structure of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed with Nadp+ and Fad in Space Group of P1211
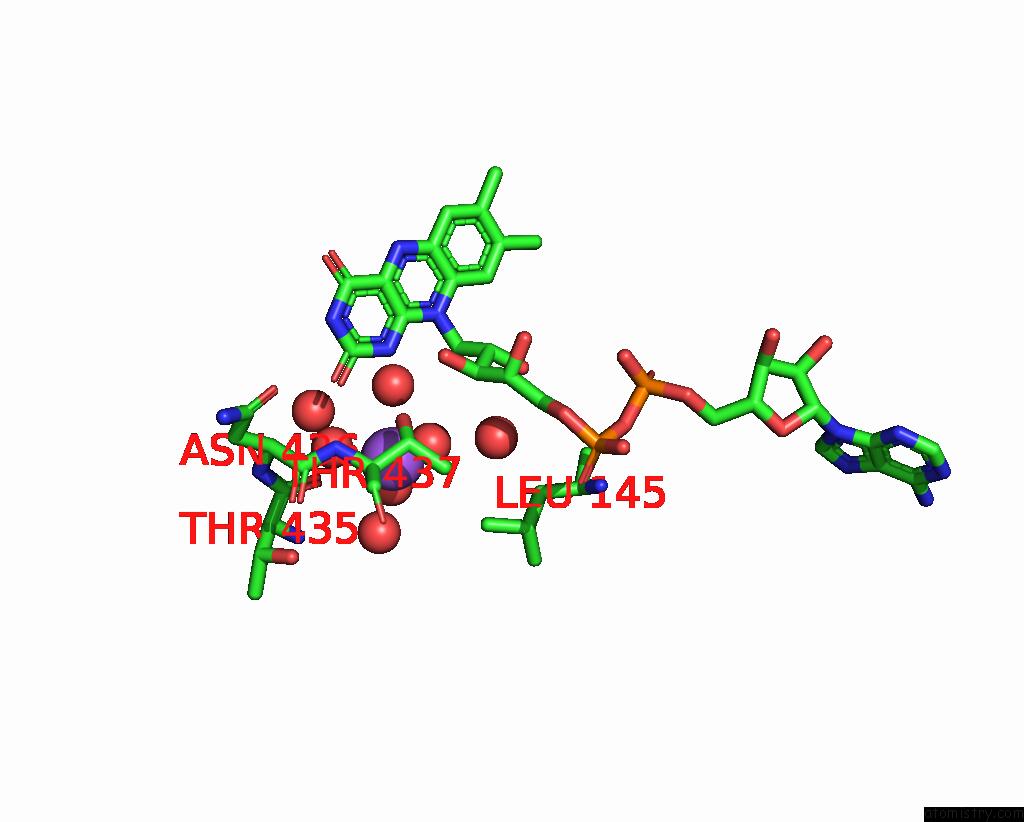
Mono view
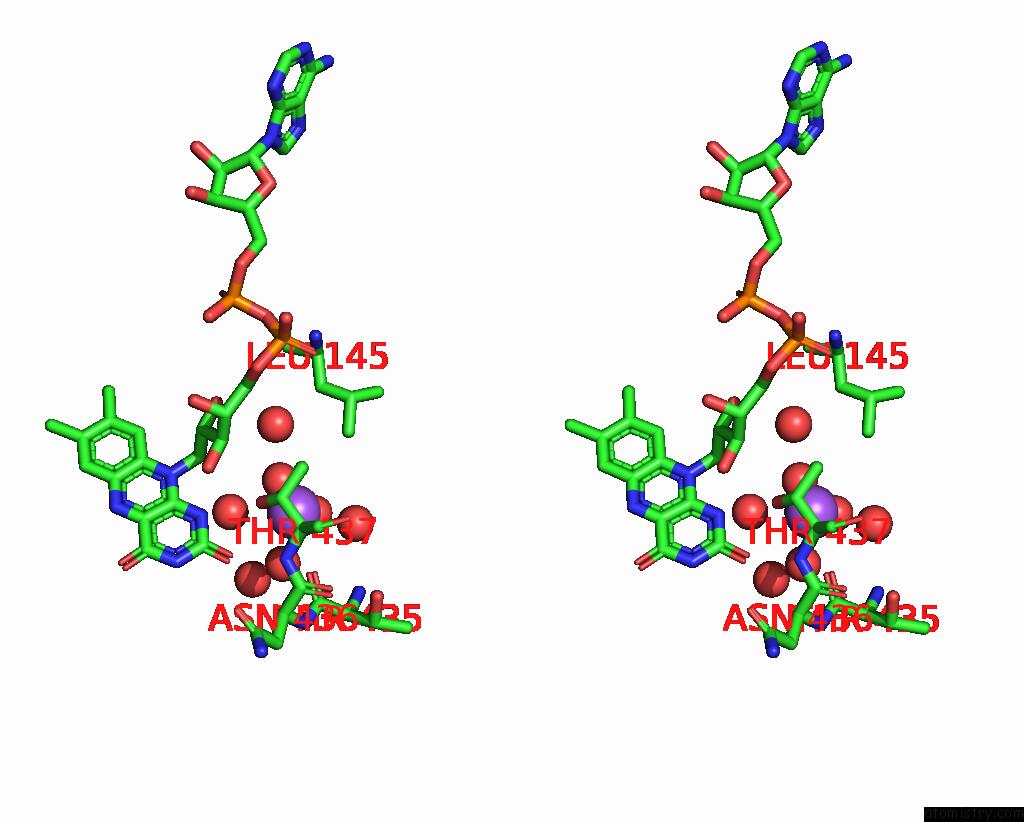
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 4 of Crystal Structure of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed with Nadp+ and Fad in Space Group of P1211 within 5.0Å range:
|
Sodium binding site 5 out of 5 in 7v8s
Go back to
Sodium binding site 5 out
of 5 in the Crystal Structure of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed with Nadp+ and Fad in Space Group of P1211
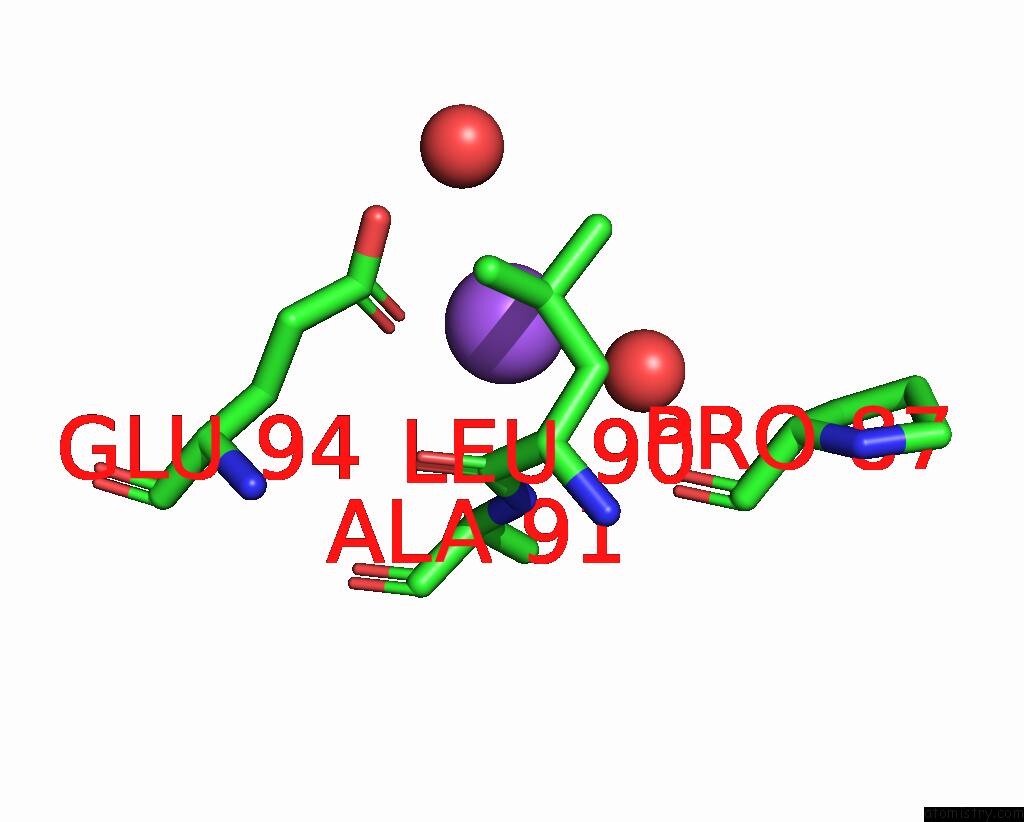
Mono view
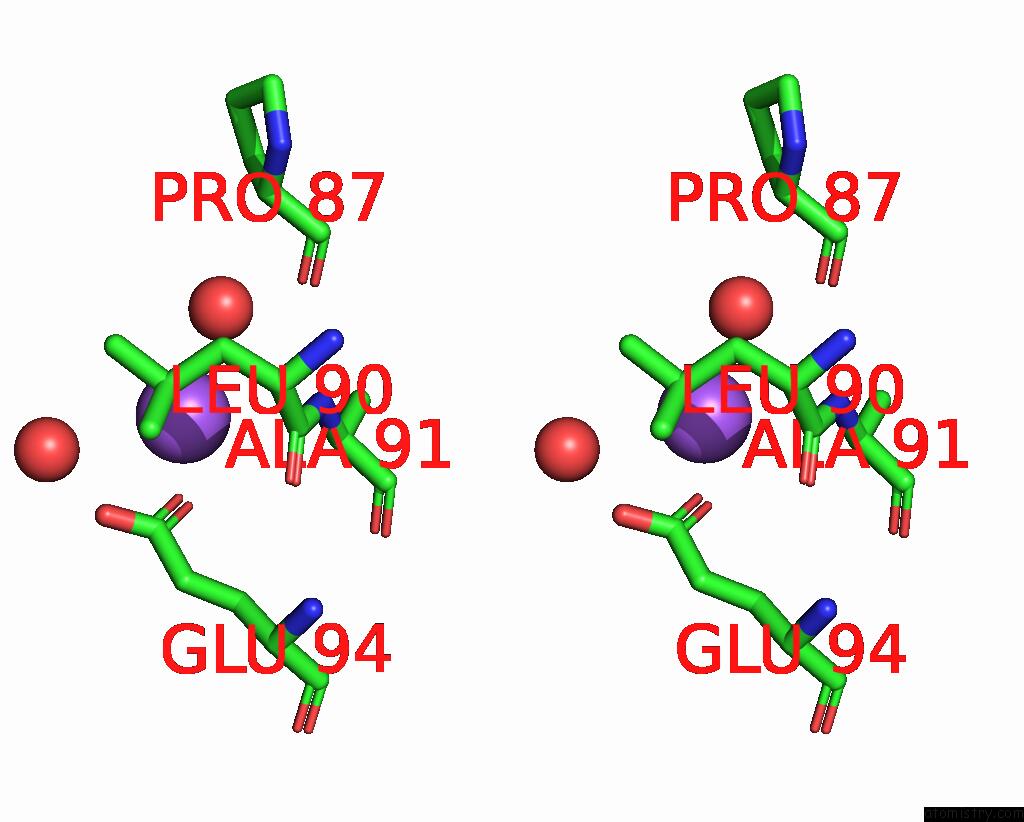
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 5 of Crystal Structure of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed with Nadp+ and Fad in Space Group of P1211 within 5.0Å range:
|
Reference:
Y.J.Dong,
T.Li,
S.Q.Zhang,
J.Sanchis,
H.Yin,
J.Ren,
X.Sheng,
G.Y.Li,
M.T.Reetz.
Biocatalytic Baeyer-Villiger Reactions: Uncovering the Source of Regioselectivity at Each Evolutionary Stage of A Mutant with Scrutiny of Fleeting Chiral Intermediates. Acs Catalysis V. 12 3669 2022.
ISSN: ESSN 2155-5435
DOI: 10.1021/ACSCATAL.2C00415
Page generated: Mon Aug 18 12:13:04 2025
ISSN: ESSN 2155-5435
DOI: 10.1021/ACSCATAL.2C00415
Last articles
Na in 8FSWNa in 8FSV
Na in 8FSU
Na in 8FST
Na in 8FSM
Na in 8FSN
Na in 8FSH
Na in 8FSG
Na in 8FSC
Na in 8FSF