Sodium »
PDB 7mz6-7nkf »
7mzd »
Sodium in PDB 7mzd: Cryo-Em Structure of Minimal TRPV1 with Rtx Bound in C2 State
Sodium Binding Sites:
The binding sites of Sodium atom in the Cryo-Em Structure of Minimal TRPV1 with Rtx Bound in C2 State
(pdb code 7mzd). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 2 binding sites of Sodium where determined in the Cryo-Em Structure of Minimal TRPV1 with Rtx Bound in C2 State, PDB code: 7mzd:
Jump to Sodium binding site number: 1; 2;
In total 2 binding sites of Sodium where determined in the Cryo-Em Structure of Minimal TRPV1 with Rtx Bound in C2 State, PDB code: 7mzd:
Jump to Sodium binding site number: 1; 2;
Sodium binding site 1 out of 2 in 7mzd
Go back to
Sodium binding site 1 out
of 2 in the Cryo-Em Structure of Minimal TRPV1 with Rtx Bound in C2 State
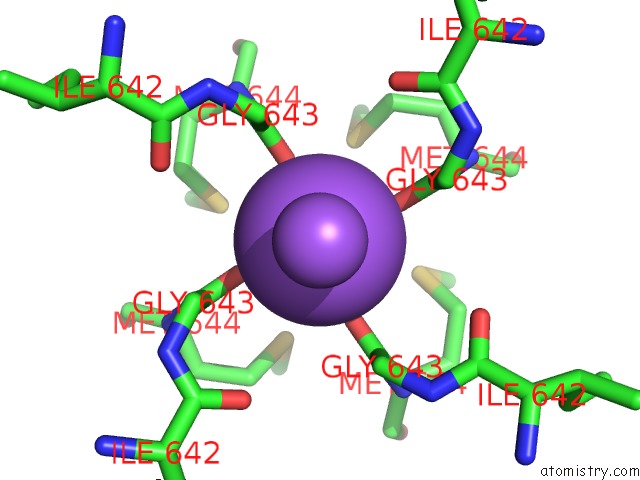
Mono view
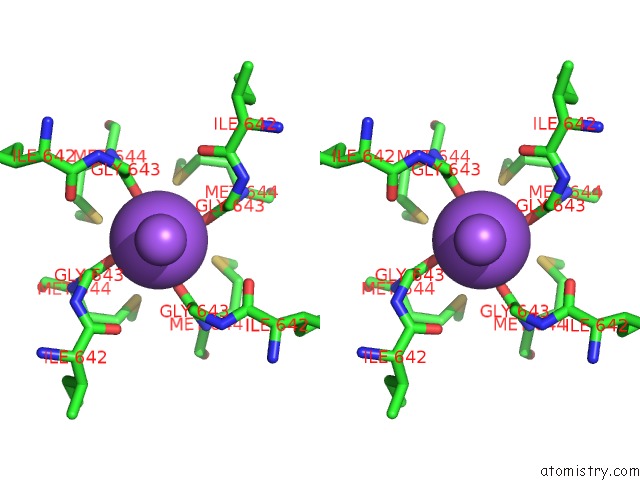
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Cryo-Em Structure of Minimal TRPV1 with Rtx Bound in C2 State within 5.0Å range:
|
Sodium binding site 2 out of 2 in 7mzd
Go back to
Sodium binding site 2 out
of 2 in the Cryo-Em Structure of Minimal TRPV1 with Rtx Bound in C2 State
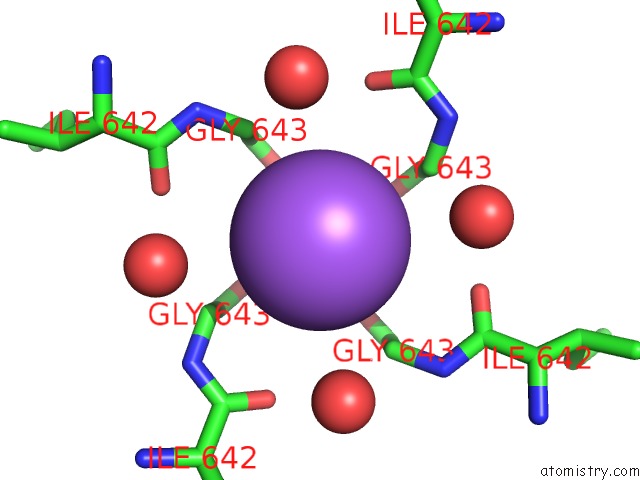
Mono view
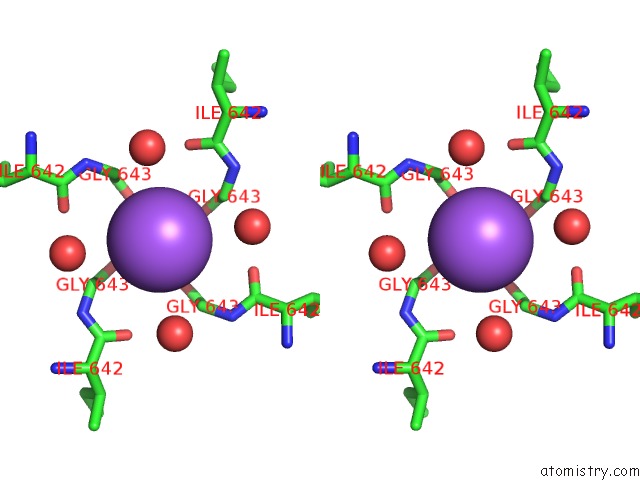
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Cryo-Em Structure of Minimal TRPV1 with Rtx Bound in C2 State within 5.0Å range:
|
Reference:
K.Zhang,
D.Julius,
Y.Cheng.
Structural Snapshots of TRPV1 Reveal Mechanism of Polymodal Functionality. Cell 2021.
ISSN: ISSN 1097-4172
PubMed: 34496225
DOI: 10.1016/J.CELL.2021.08.012
Page generated: Mon Aug 18 11:01:58 2025
ISSN: ISSN 1097-4172
PubMed: 34496225
DOI: 10.1016/J.CELL.2021.08.012
Last articles
Na in 8BAINa in 8B9Z
Na in 8B6E
Na in 8B9T
Na in 8B6D
Na in 8B5D
Na in 8B5M
Na in 8B5E
Na in 8B45
Na in 8B4X