Sodium »
PDB 6u6u-6v04 »
6uao »
Sodium in PDB 6uao: Imidazole-Triggered Ras-Specific Subtilisin SUBT_BACAM Complexed with the Peptide Eeysam
Protein crystallography data
The structure of Imidazole-Triggered Ras-Specific Subtilisin SUBT_BACAM Complexed with the Peptide Eeysam, PDB code: 6uao
was solved by
E.A.Toth,
P.N.Bryan,
J.Orban,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 41.41 / 1.63 |
| Space group | P 41 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 58.510, 58.510, 125.090, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 14.9 / 18 |
Other elements in 6uao:
The structure of Imidazole-Triggered Ras-Specific Subtilisin SUBT_BACAM Complexed with the Peptide Eeysam also contains other interesting chemical elements:
| Chlorine | (Cl) | 1 atom |
Sodium Binding Sites:
The binding sites of Sodium atom in the Imidazole-Triggered Ras-Specific Subtilisin SUBT_BACAM Complexed with the Peptide Eeysam
(pdb code 6uao). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Imidazole-Triggered Ras-Specific Subtilisin SUBT_BACAM Complexed with the Peptide Eeysam, PDB code: 6uao:
In total only one binding site of Sodium was determined in the Imidazole-Triggered Ras-Specific Subtilisin SUBT_BACAM Complexed with the Peptide Eeysam, PDB code: 6uao:
Sodium binding site 1 out of 1 in 6uao
Go back to
Sodium binding site 1 out
of 1 in the Imidazole-Triggered Ras-Specific Subtilisin SUBT_BACAM Complexed with the Peptide Eeysam
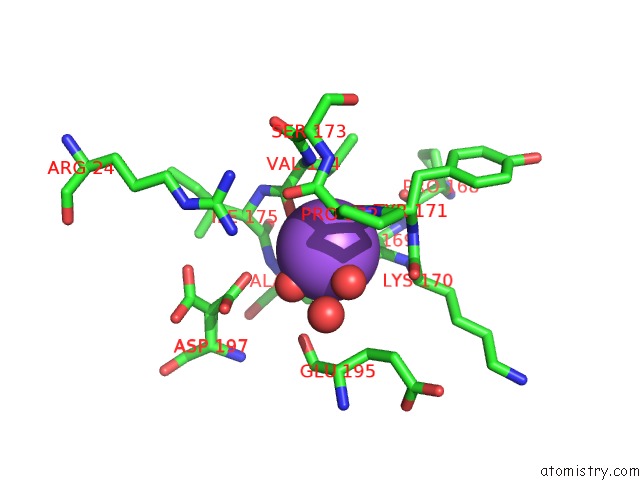
Mono view
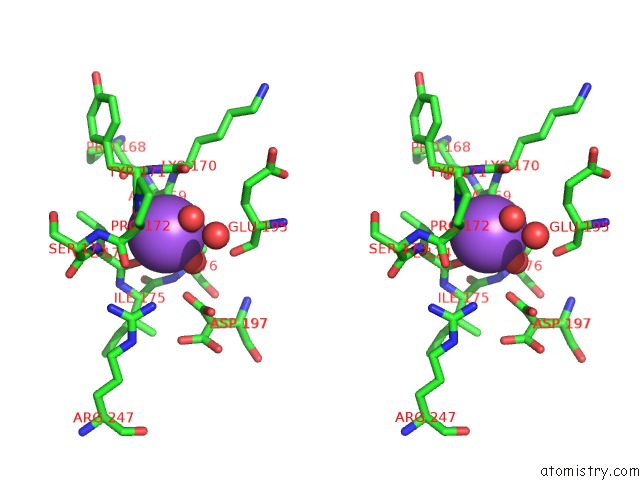
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Imidazole-Triggered Ras-Specific Subtilisin SUBT_BACAM Complexed with the Peptide Eeysam within 5.0Å range:
|
Reference:
E.A.Toth,
P.N.Bryan.
A Programmable Proteolytic Machine That Specifically Degrades Oncogenic Ras in Vitro, in Engineered Bacteria, and in Human Cell Culture To Be Published.
Page generated: Tue Oct 8 14:08:53 2024
Last articles
F in 4GE2F in 4G9C
F in 4GAB
F in 4GA8
F in 4G90
F in 4G8R
F in 4G7G
F in 4G8O
F in 4G6O
F in 4G2R