Sodium »
PDB 6d50-6dmi »
6dmc »
Sodium in PDB 6dmc: Ppgpp Riboswitch Bound to Ppgpp, Native Structure
Protein crystallography data
The structure of Ppgpp Riboswitch Bound to Ppgpp, Native Structure, PDB code: 6dmc
was solved by
A.Peselis,
A.Serganov,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.00 / 2.20 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 78.162, 48.917, 78.723, 90.00, 97.33, 90.00 |
| R / Rfree (%) | 22.8 / 25.9 |
Other elements in 6dmc:
The structure of Ppgpp Riboswitch Bound to Ppgpp, Native Structure also contains other interesting chemical elements:
| Magnesium | (Mg) | 21 atoms |
| Potassium | (K) | 1 atom |
Sodium Binding Sites:
The binding sites of Sodium atom in the Ppgpp Riboswitch Bound to Ppgpp, Native Structure
(pdb code 6dmc). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 10 binding sites of Sodium where determined in the Ppgpp Riboswitch Bound to Ppgpp, Native Structure, PDB code: 6dmc:
Jump to Sodium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
In total 10 binding sites of Sodium where determined in the Ppgpp Riboswitch Bound to Ppgpp, Native Structure, PDB code: 6dmc:
Jump to Sodium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Sodium binding site 1 out of 10 in 6dmc
Go back to
Sodium binding site 1 out
of 10 in the Ppgpp Riboswitch Bound to Ppgpp, Native Structure
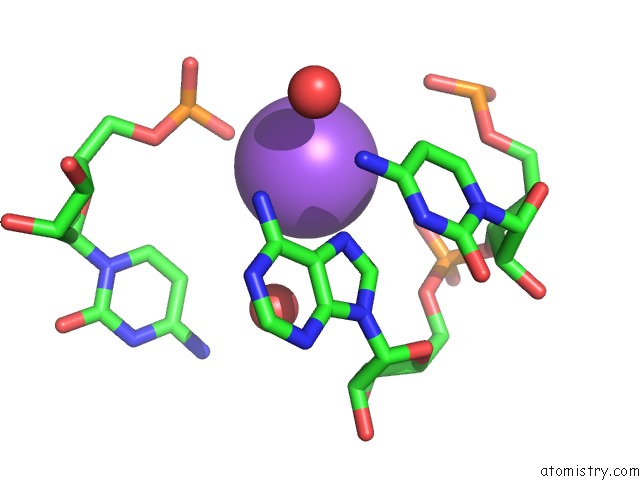
Mono view
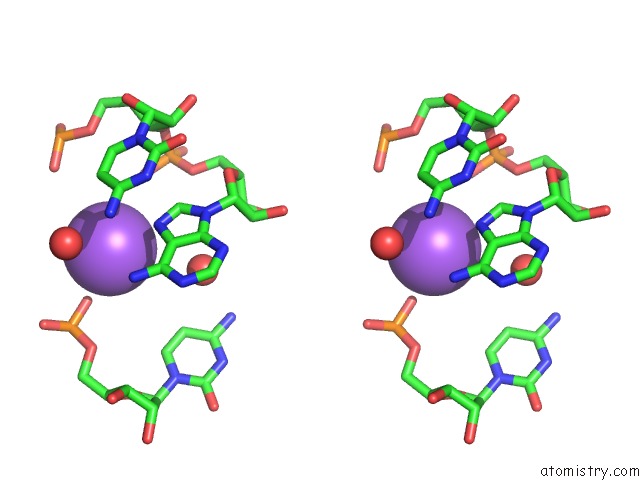
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Ppgpp Riboswitch Bound to Ppgpp, Native Structure within 5.0Å range:
|
Sodium binding site 2 out of 10 in 6dmc
Go back to
Sodium binding site 2 out
of 10 in the Ppgpp Riboswitch Bound to Ppgpp, Native Structure
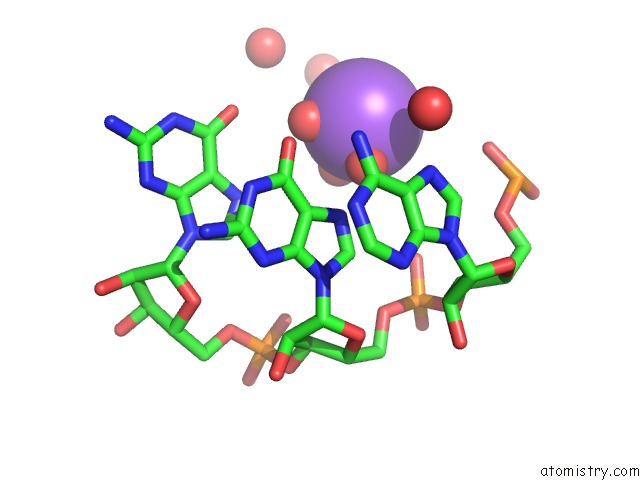
Mono view
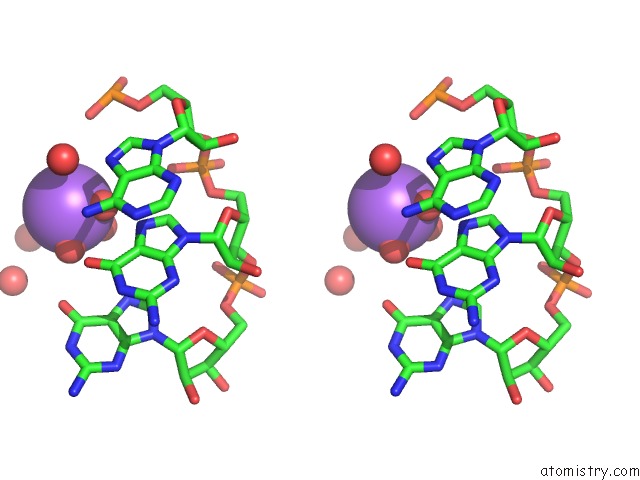
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Ppgpp Riboswitch Bound to Ppgpp, Native Structure within 5.0Å range:
|
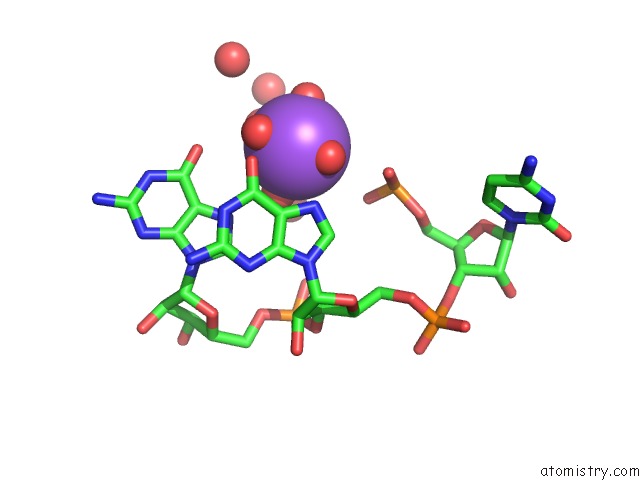
Sodium binding site 3 out of 10 in 6dmc
Go back to
Sodium binding site 3 out
of 10 in the Ppgpp Riboswitch Bound to Ppgpp, Native Structure
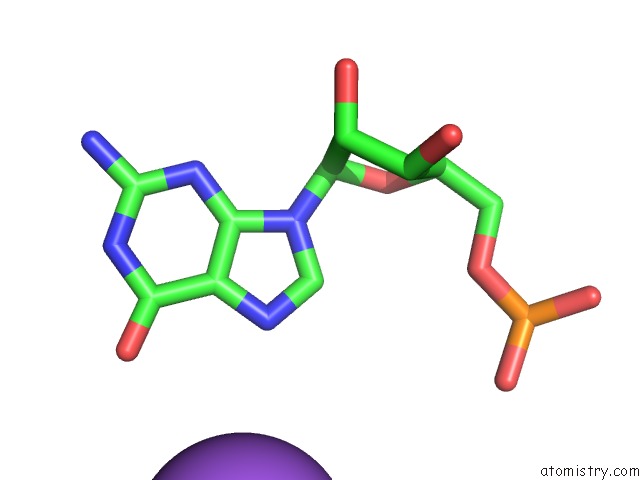
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 3 of Ppgpp Riboswitch Bound to Ppgpp, Native Structure within 5.0Å range:
|
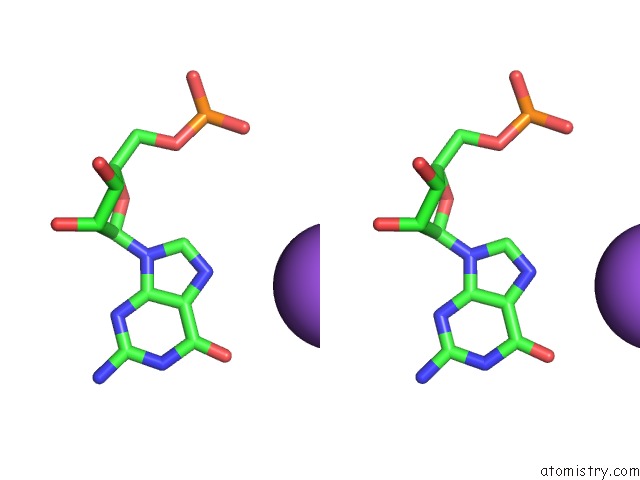
Sodium binding site 4 out of 10 in 6dmc
Go back to
Sodium binding site 4 out
of 10 in the Ppgpp Riboswitch Bound to Ppgpp, Native Structure
Mono view
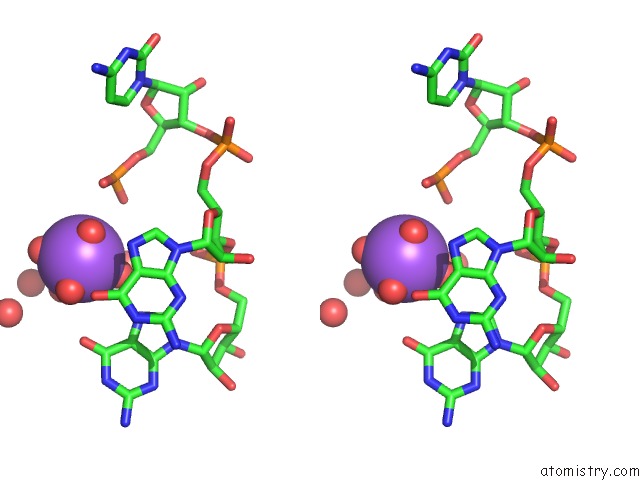
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 4 of Ppgpp Riboswitch Bound to Ppgpp, Native Structure within 5.0Å range:
|
Sodium binding site 5 out of 10 in 6dmc
Go back to
Sodium binding site 5 out
of 10 in the Ppgpp Riboswitch Bound to Ppgpp, Native Structure
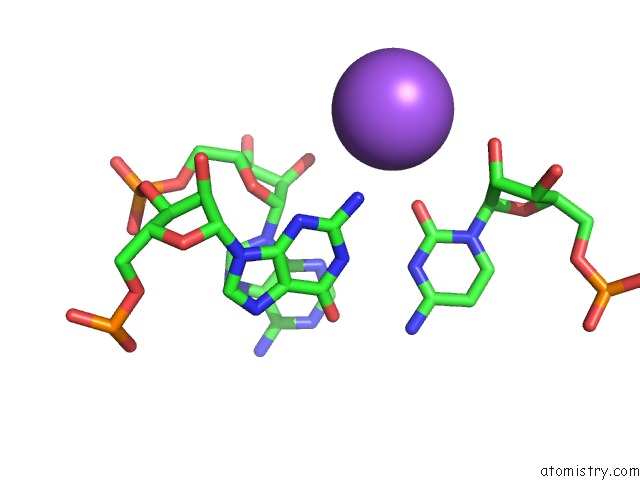
Mono view
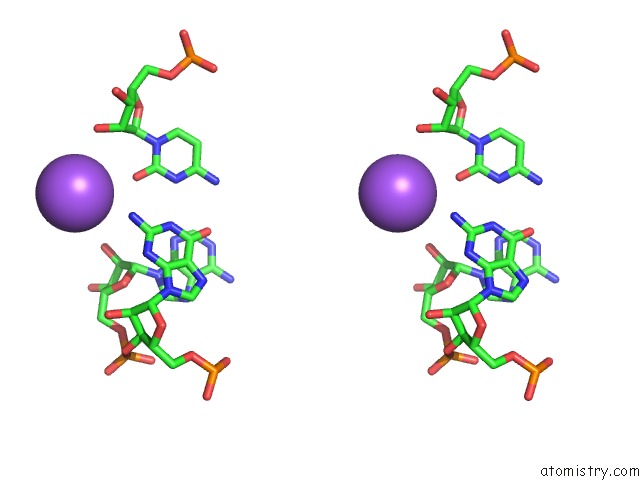
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 5 of Ppgpp Riboswitch Bound to Ppgpp, Native Structure within 5.0Å range:
|
Sodium binding site 6 out of 10 in 6dmc
Go back to
Sodium binding site 6 out
of 10 in the Ppgpp Riboswitch Bound to Ppgpp, Native Structure
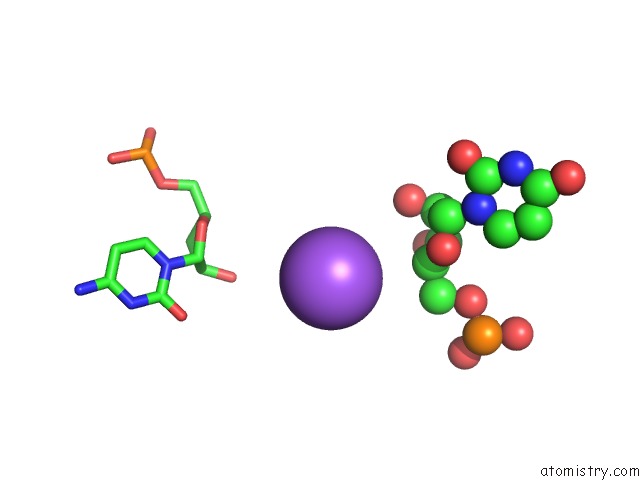
Mono view
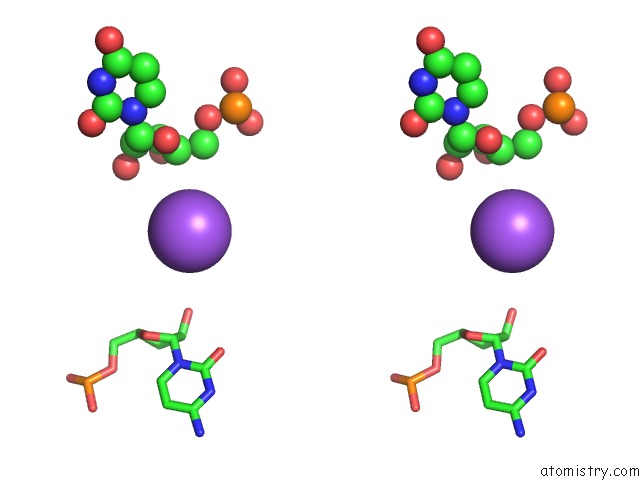
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 6 of Ppgpp Riboswitch Bound to Ppgpp, Native Structure within 5.0Å range:
|
Sodium binding site 7 out of 10 in 6dmc
Go back to
Sodium binding site 7 out
of 10 in the Ppgpp Riboswitch Bound to Ppgpp, Native Structure
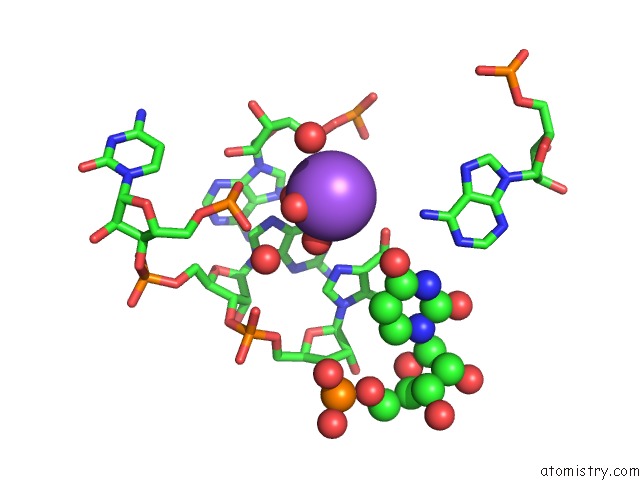
Mono view
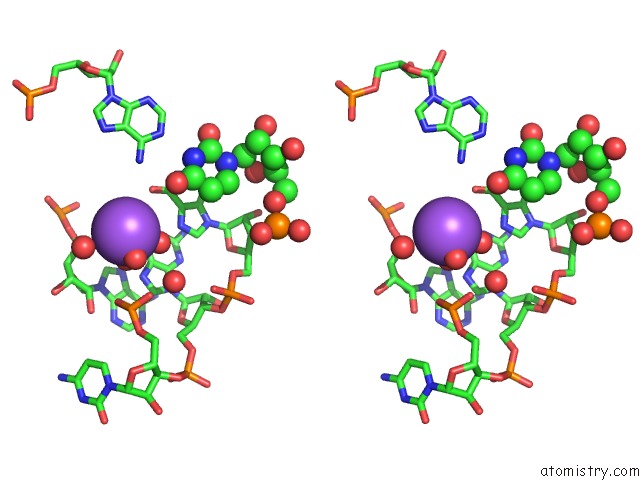
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 7 of Ppgpp Riboswitch Bound to Ppgpp, Native Structure within 5.0Å range:
|
Sodium binding site 8 out of 10 in 6dmc
Go back to
Sodium binding site 8 out
of 10 in the Ppgpp Riboswitch Bound to Ppgpp, Native Structure
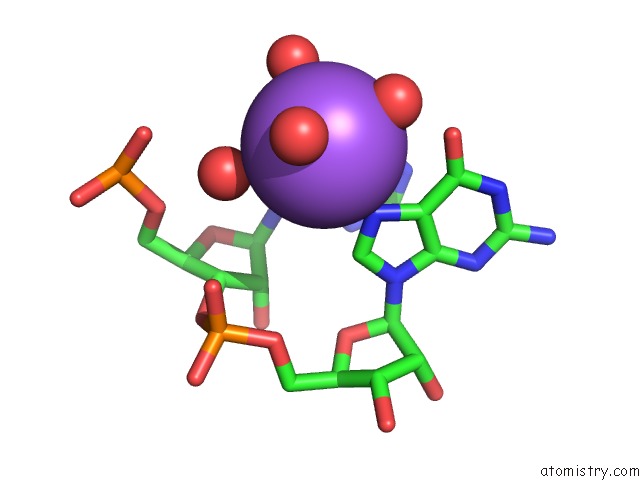
Mono view
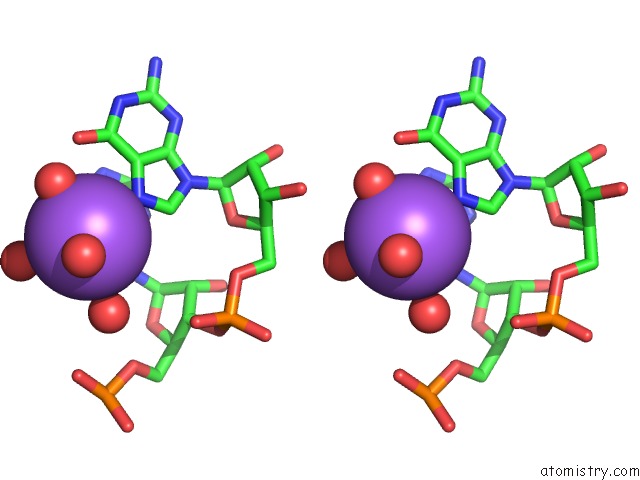
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 8 of Ppgpp Riboswitch Bound to Ppgpp, Native Structure within 5.0Å range:
|
Sodium binding site 9 out of 10 in 6dmc
Go back to
Sodium binding site 9 out
of 10 in the Ppgpp Riboswitch Bound to Ppgpp, Native Structure
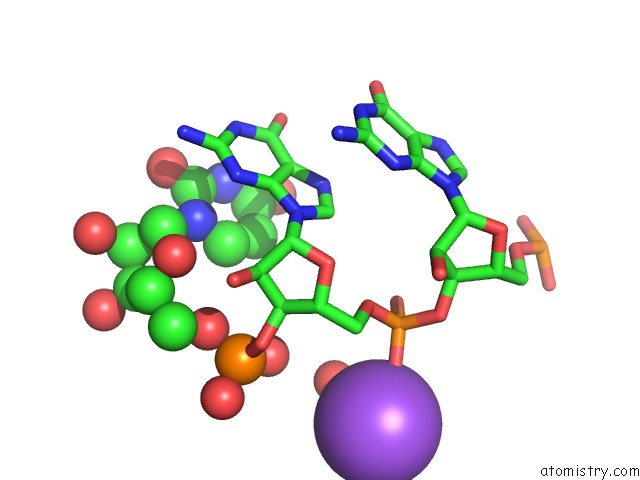
Mono view
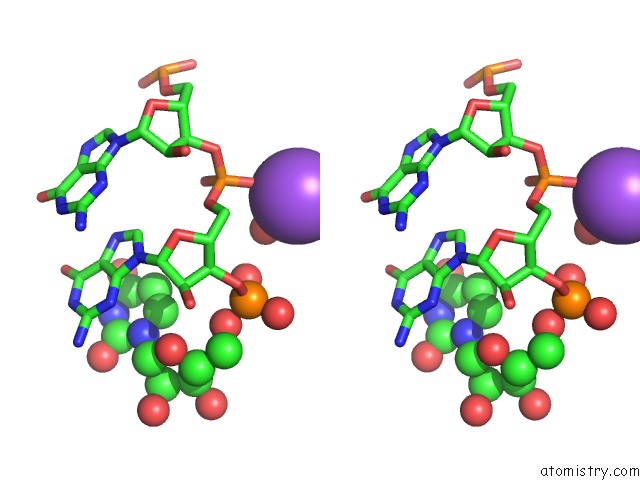
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 9 of Ppgpp Riboswitch Bound to Ppgpp, Native Structure within 5.0Å range:
|
Sodium binding site 10 out of 10 in 6dmc
Go back to
Sodium binding site 10 out
of 10 in the Ppgpp Riboswitch Bound to Ppgpp, Native Structure
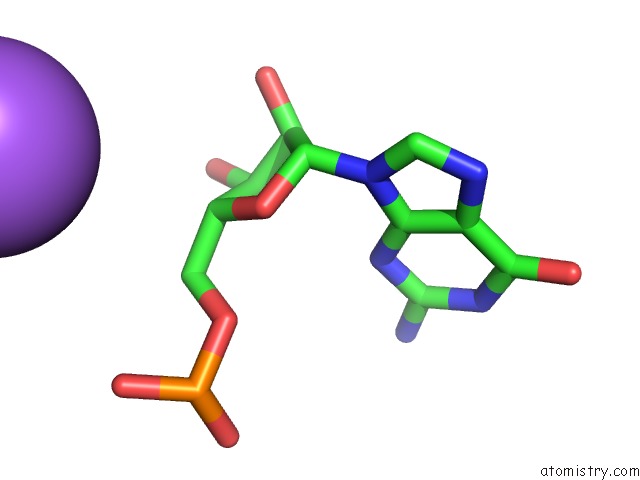
Mono view
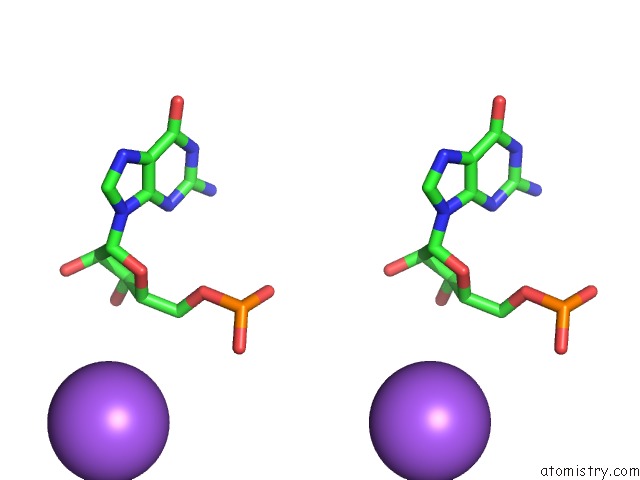
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 10 of Ppgpp Riboswitch Bound to Ppgpp, Native Structure within 5.0Å range:
|
Reference:
A.Peselis,
A.Serganov.
Ykkc Riboswitches Employ An Add-on Helix to Adjust Specificity For Polyanionic Ligands. Nat. Chem. Biol. V. 14 887 2018.
ISSN: ESSN 1552-4469
PubMed: 30120360
DOI: 10.1038/S41589-018-0114-4
Page generated: Tue Oct 8 07:44:20 2024
ISSN: ESSN 1552-4469
PubMed: 30120360
DOI: 10.1038/S41589-018-0114-4
Last articles
Fe in 2YXOFe in 2YRS
Fe in 2YXC
Fe in 2YNM
Fe in 2YVJ
Fe in 2YP1
Fe in 2YU2
Fe in 2YU1
Fe in 2YQB
Fe in 2YOO