Sodium »
PDB 6d50-6dmi »
6d7a »
Sodium in PDB 6d7a: Structure of T. Gondii PLP1 Beta-Rich Domain
Protein crystallography data
The structure of Structure of T. Gondii PLP1 Beta-Rich Domain, PDB code: 6d7a
was solved by
A.J.Guerra,
N.M.Koropatkin,
Z.Wawrzak,
C.M.E.Bahr,
V.B.Carruthers,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 24.88 / 1.13 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 101.980, 50.850, 105.340, 90.00, 90.13, 90.00 |
| R / Rfree (%) | 14.1 / 16.4 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Structure of T. Gondii PLP1 Beta-Rich Domain
(pdb code 6d7a). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 4 binding sites of Sodium where determined in the Structure of T. Gondii PLP1 Beta-Rich Domain, PDB code: 6d7a:
Jump to Sodium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Sodium where determined in the Structure of T. Gondii PLP1 Beta-Rich Domain, PDB code: 6d7a:
Jump to Sodium binding site number: 1; 2; 3; 4;
Sodium binding site 1 out of 4 in 6d7a
Go back to
Sodium binding site 1 out
of 4 in the Structure of T. Gondii PLP1 Beta-Rich Domain
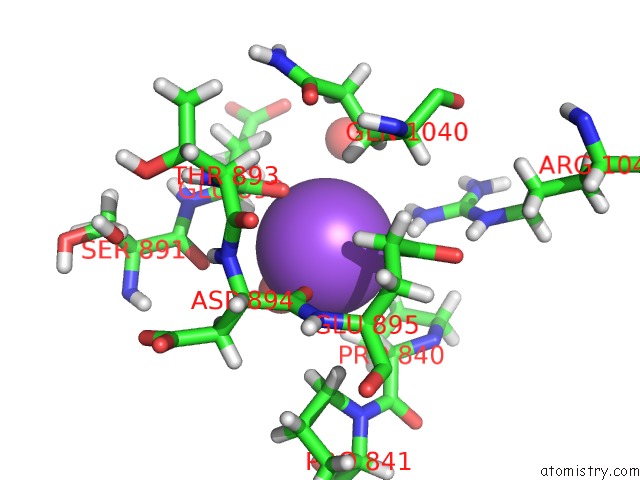
Mono view
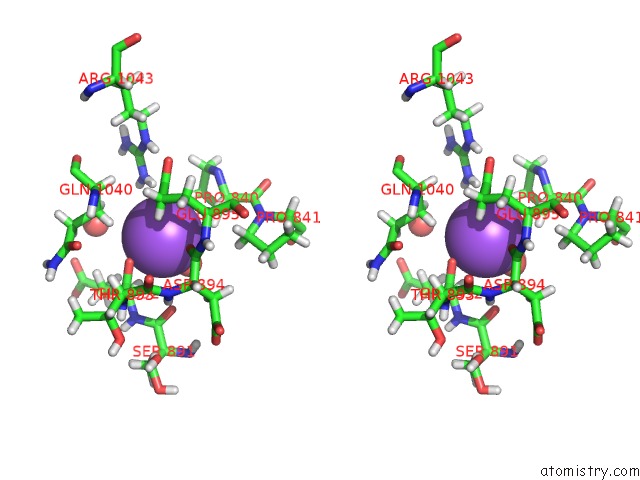
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Structure of T. Gondii PLP1 Beta-Rich Domain within 5.0Å range:
|
Sodium binding site 2 out of 4 in 6d7a
Go back to
Sodium binding site 2 out
of 4 in the Structure of T. Gondii PLP1 Beta-Rich Domain
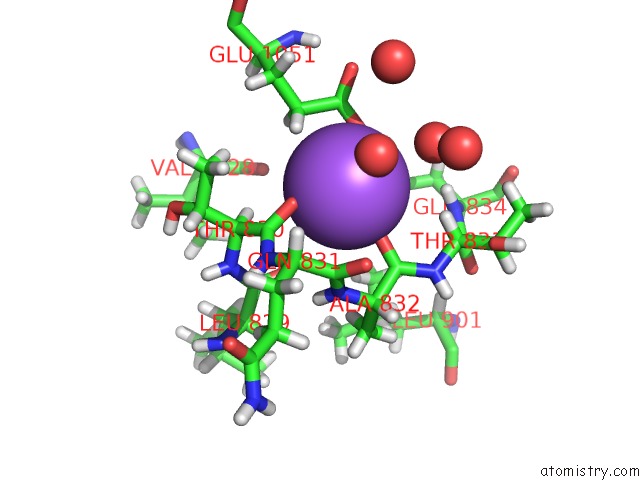
Mono view
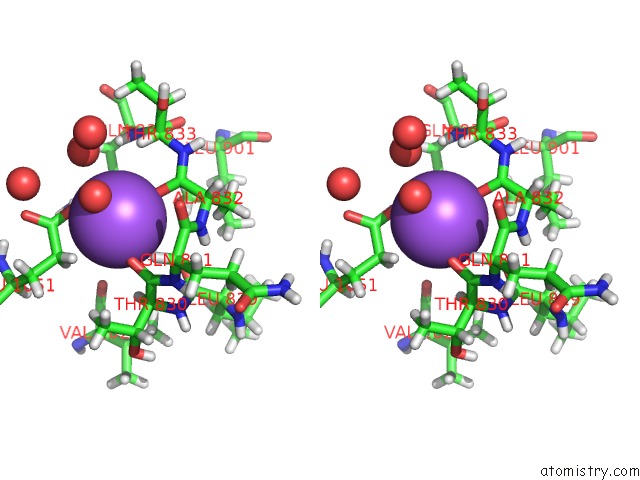
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Structure of T. Gondii PLP1 Beta-Rich Domain within 5.0Å range:
|
Sodium binding site 3 out of 4 in 6d7a
Go back to
Sodium binding site 3 out
of 4 in the Structure of T. Gondii PLP1 Beta-Rich Domain
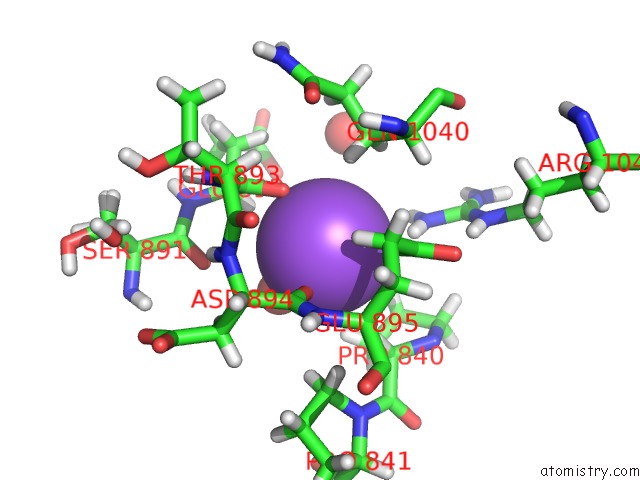
Mono view
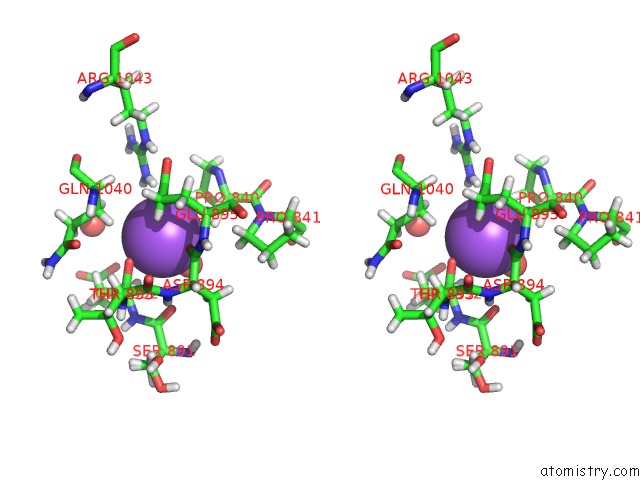
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 3 of Structure of T. Gondii PLP1 Beta-Rich Domain within 5.0Å range:
|
Sodium binding site 4 out of 4 in 6d7a
Go back to
Sodium binding site 4 out
of 4 in the Structure of T. Gondii PLP1 Beta-Rich Domain
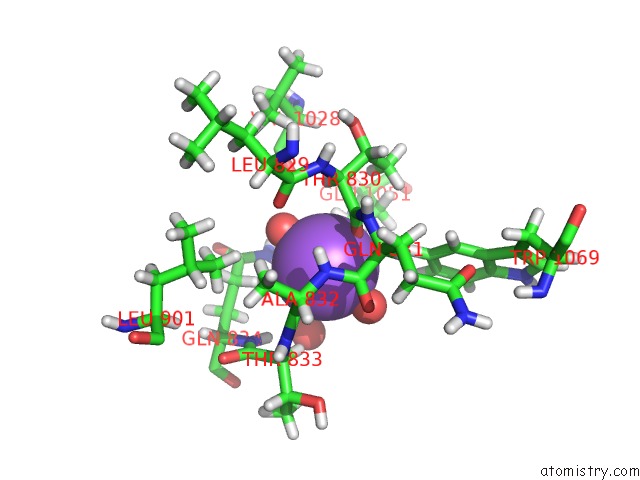
Mono view
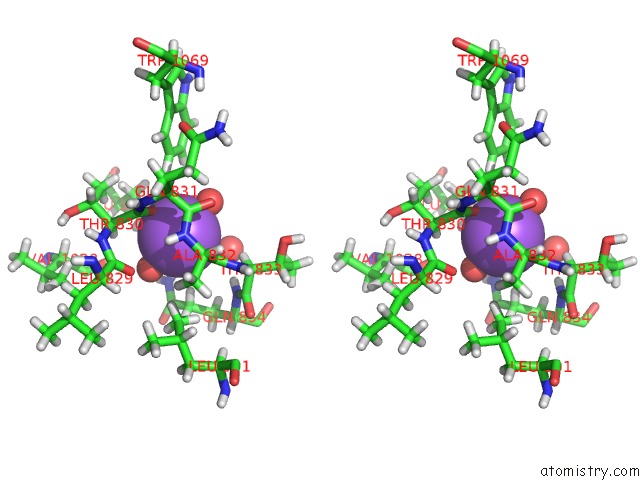
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 4 of Structure of T. Gondii PLP1 Beta-Rich Domain within 5.0Å range:
|
Reference:
A.J.Guerra,
O.Zhang,
C.M.E.Bahr,
M.H.Huynh,
J.Delproposto,
W.C.Brown,
Z.Wawrzak,
N.M.Koropatkin,
V.B.Carruthers.
Structural Basis of Toxoplasma Gondii Perforin-Like Protein 1 Membrane Interaction and Activity During Egress. Plos Pathog. V. 14 07476 2018.
ISSN: ESSN 1553-7374
PubMed: 30513119
DOI: 10.1371/JOURNAL.PPAT.1007476
Page generated: Tue Oct 8 07:41:37 2024
ISSN: ESSN 1553-7374
PubMed: 30513119
DOI: 10.1371/JOURNAL.PPAT.1007476
Last articles
Ca in 5M8ECa in 5M7G
Ca in 5M6C
Ca in 5M7E
Ca in 5M69
Ca in 5M6D
Ca in 5M5F
Ca in 5M3C
Ca in 5M2S
Ca in 5M62