Sodium »
PDB 6c12-6chk »
6c5b »
Sodium in PDB 6c5b: Crystal Structure Analysis of Laphzm
Protein crystallography data
The structure of Crystal Structure Analysis of Laphzm, PDB code: 6c5b
was solved by
D.G.Beltran,
A.Schacht,
L.Zhang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 71.42 / 1.42 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 56.275, 81.017, 72.164, 90.00, 98.23, 90.00 |
| R / Rfree (%) | 18.1 / 20.3 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Crystal Structure Analysis of Laphzm
(pdb code 6c5b). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 3 binding sites of Sodium where determined in the Crystal Structure Analysis of Laphzm, PDB code: 6c5b:
Jump to Sodium binding site number: 1; 2; 3;
In total 3 binding sites of Sodium where determined in the Crystal Structure Analysis of Laphzm, PDB code: 6c5b:
Jump to Sodium binding site number: 1; 2; 3;
Sodium binding site 1 out of 3 in 6c5b
Go back to
Sodium binding site 1 out
of 3 in the Crystal Structure Analysis of Laphzm
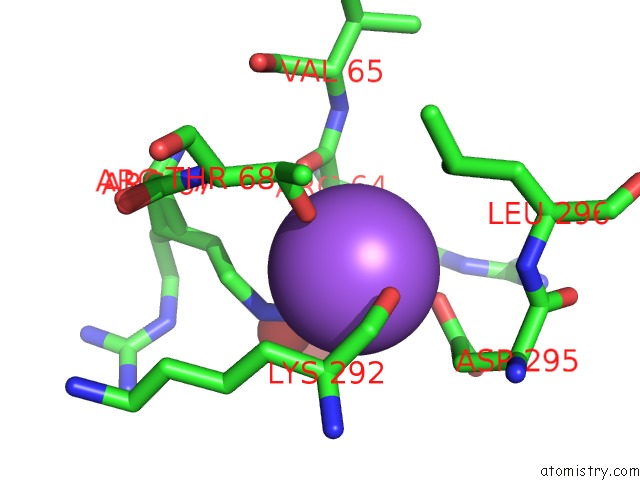
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Crystal Structure Analysis of Laphzm within 5.0Å range:
|
Sodium binding site 2 out of 3 in 6c5b
Go back to
Sodium binding site 2 out
of 3 in the Crystal Structure Analysis of Laphzm

Mono view
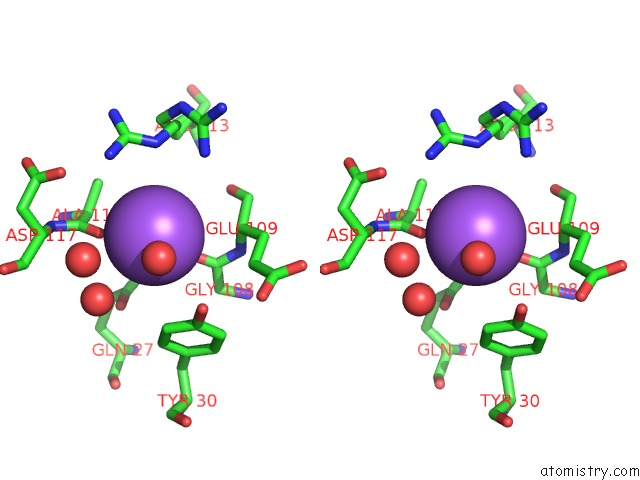
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Crystal Structure Analysis of Laphzm within 5.0Å range:
|
Sodium binding site 3 out of 3 in 6c5b
Go back to
Sodium binding site 3 out
of 3 in the Crystal Structure Analysis of Laphzm

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 3 of Crystal Structure Analysis of Laphzm within 5.0Å range:
|
Reference:
J.Jiang,
D.Guiza Beltran,
A.Schacht,
S.Wright,
L.Zhang,
L.Du.
Functional and Structural Analysis of Phenazine O-Methyltransferase Laphzm From Lysobacter Antibioticus OH13 and One-Pot Enzymatic Synthesis of the Antibiotic Myxin. Acs Chem. Biol. V. 13 1003 2018.
ISSN: ESSN 1554-8937
PubMed: 29510028
DOI: 10.1021/ACSCHEMBIO.8B00062
Page generated: Tue Oct 8 06:30:31 2024
ISSN: ESSN 1554-8937
PubMed: 29510028
DOI: 10.1021/ACSCHEMBIO.8B00062
Last articles
Cl in 5GIZCl in 5GG6
Cl in 5GGY
Cl in 5GHU
Cl in 5G6Q
Cl in 5G6P
Cl in 5G6O
Cl in 5G6L
Cl in 5G6N
Cl in 5G6M