Sodium »
PDB 5hn3-5i9b »
5i8u »
Sodium in PDB 5i8u: Crystal Structure of the RV1700 (Mt Adprase) E142Q Mutant
Enzymatic activity of Crystal Structure of the RV1700 (Mt Adprase) E142Q Mutant
All present enzymatic activity of Crystal Structure of the RV1700 (Mt Adprase) E142Q Mutant:
3.6.1.13;
3.6.1.13;
Protein crystallography data
The structure of Crystal Structure of the RV1700 (Mt Adprase) E142Q Mutant, PDB code: 5i8u
was solved by
P.Thirawatananond,
L.-W.Kang,
L.M.Amzel,
S.B.Gabelli,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 141.42 / 2.00 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 152.768, 124.844, 142.959, 90.00, 98.50, 90.00 |
| R / Rfree (%) | 22.2 / 25.8 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Crystal Structure of the RV1700 (Mt Adprase) E142Q Mutant
(pdb code 5i8u). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 2 binding sites of Sodium where determined in the Crystal Structure of the RV1700 (Mt Adprase) E142Q Mutant, PDB code: 5i8u:
Jump to Sodium binding site number: 1; 2;
In total 2 binding sites of Sodium where determined in the Crystal Structure of the RV1700 (Mt Adprase) E142Q Mutant, PDB code: 5i8u:
Jump to Sodium binding site number: 1; 2;
Sodium binding site 1 out of 2 in 5i8u
Go back to
Sodium binding site 1 out
of 2 in the Crystal Structure of the RV1700 (Mt Adprase) E142Q Mutant
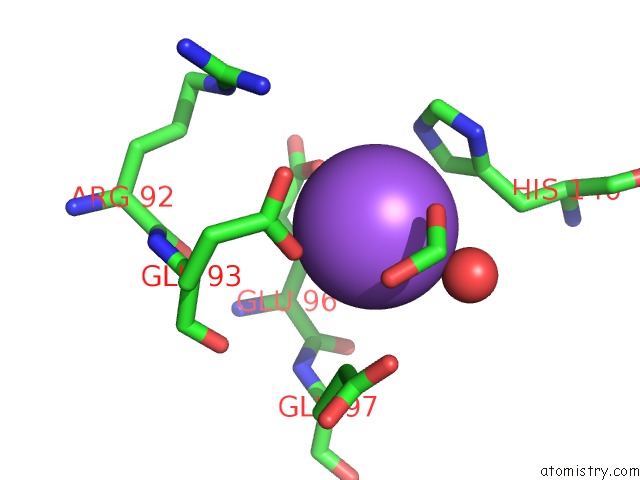
Mono view
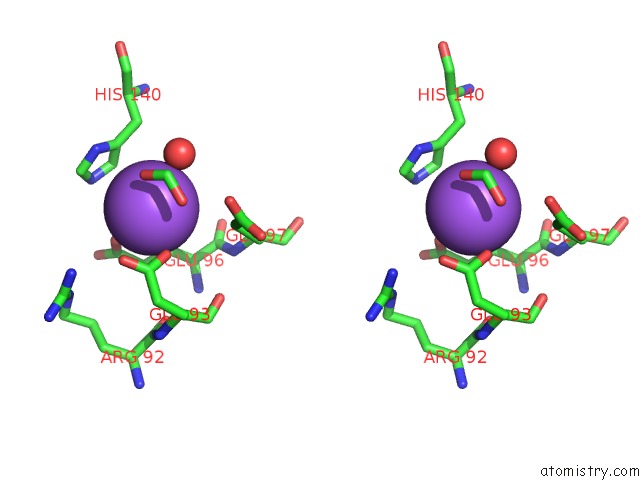
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Crystal Structure of the RV1700 (Mt Adprase) E142Q Mutant within 5.0Å range:
|
Sodium binding site 2 out of 2 in 5i8u
Go back to
Sodium binding site 2 out
of 2 in the Crystal Structure of the RV1700 (Mt Adprase) E142Q Mutant
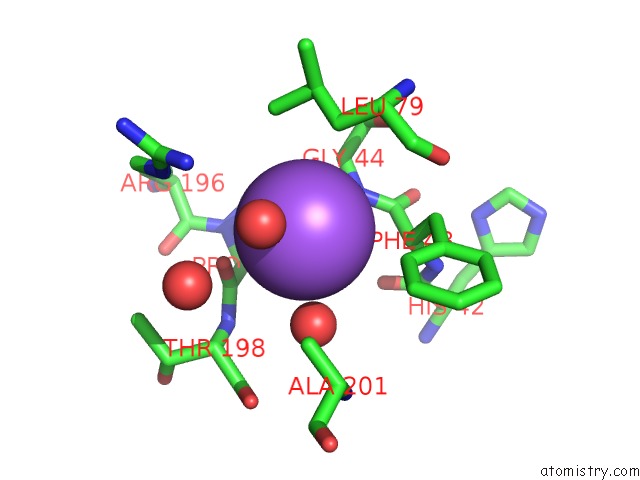
Mono view
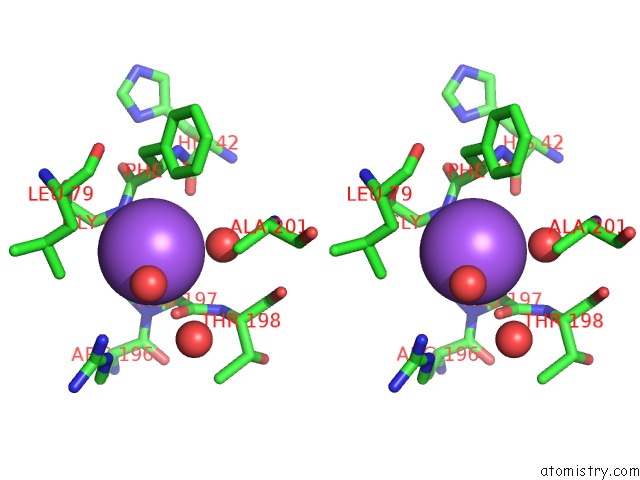
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Crystal Structure of the RV1700 (Mt Adprase) E142Q Mutant within 5.0Å range:
|
Reference:
S.F.O'handley,
P.Thirawatananond,
L.W.Kang,
J.E.Cunningham,
J.A.Leyva,
L.M.Amzel,
S.B.Gabelli.
Kinetic and Mutational Studies of the Adenosine Diphosphate Ribose Hydrolase From Mycobacterium Tuberculosis. J. Bioenerg. Biomembr. V. 48 557 2016.
ISSN: ISSN 1573-6881
PubMed: 27683242
DOI: 10.1007/S10863-016-9681-9
Page generated: Mon Aug 18 00:13:46 2025
ISSN: ISSN 1573-6881
PubMed: 27683242
DOI: 10.1007/S10863-016-9681-9
Last articles
Na in 7AAWNa in 7A6Q
Na in 7A74
Na in 7A70
Na in 7A56
Na in 7A3K
Na in 7A68
Na in 7A3I
Na in 7A5Z
Na in 7A2N