Sodium »
PDB 4zm0-5ac7 »
5a1a »
Sodium in PDB 5a1a: 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with A Cell-Permeant Inhibitor
Enzymatic activity of 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with A Cell-Permeant Inhibitor
All present enzymatic activity of 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with A Cell-Permeant Inhibitor:
3.2.1.23;
3.2.1.23;
Other elements in 5a1a:
The structure of 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with A Cell-Permeant Inhibitor also contains other interesting chemical elements:
| Magnesium | (Mg) | 8 atoms |
Sodium Binding Sites:
The binding sites of Sodium atom in the 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with A Cell-Permeant Inhibitor
(pdb code 5a1a). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 8 binding sites of Sodium where determined in the 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with A Cell-Permeant Inhibitor, PDB code: 5a1a:
Jump to Sodium binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Sodium where determined in the 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with A Cell-Permeant Inhibitor, PDB code: 5a1a:
Jump to Sodium binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Sodium binding site 1 out of 8 in 5a1a
Go back to
Sodium binding site 1 out
of 8 in the 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with A Cell-Permeant Inhibitor
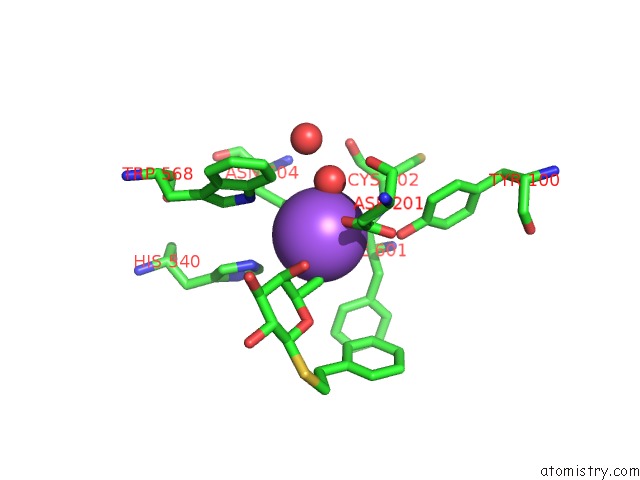
Mono view
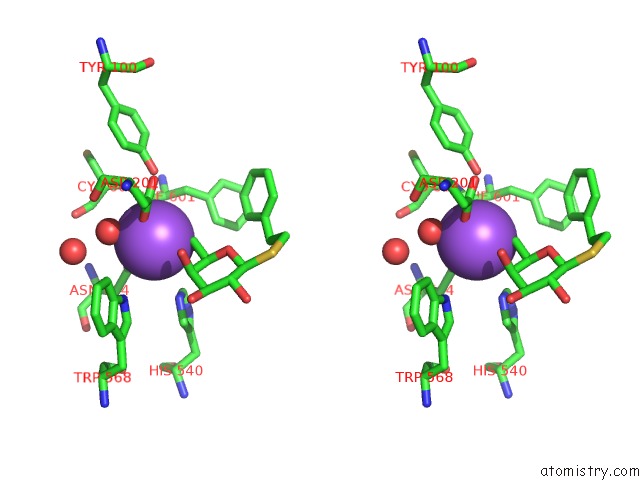
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with A Cell-Permeant Inhibitor within 5.0Å range:
|
Sodium binding site 2 out of 8 in 5a1a
Go back to
Sodium binding site 2 out
of 8 in the 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with A Cell-Permeant Inhibitor
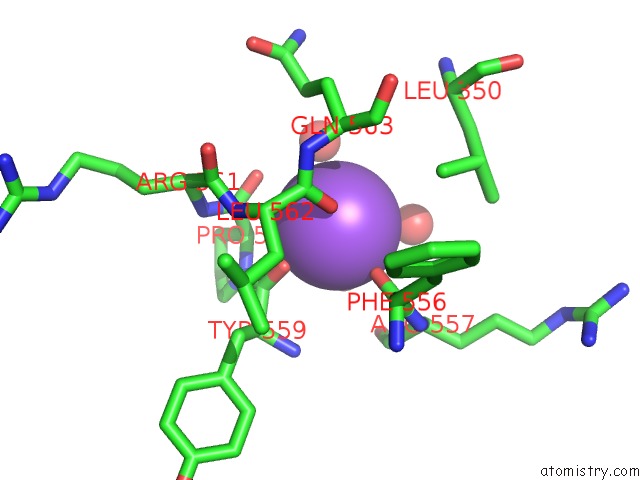
Mono view
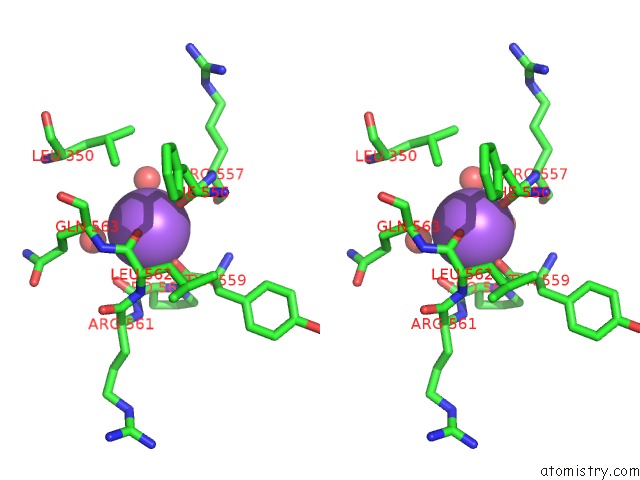
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with A Cell-Permeant Inhibitor within 5.0Å range:
|
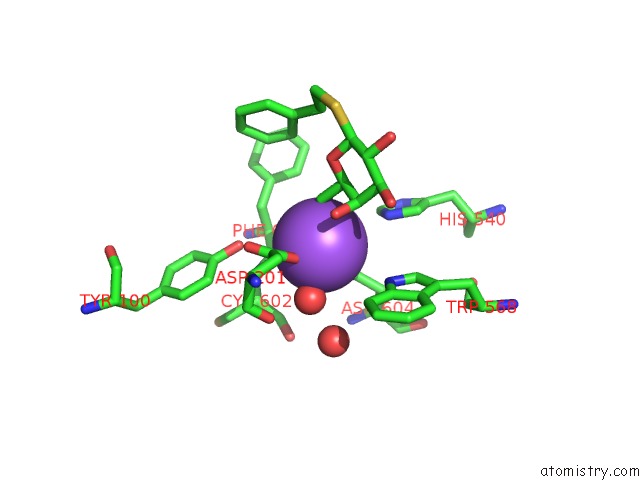
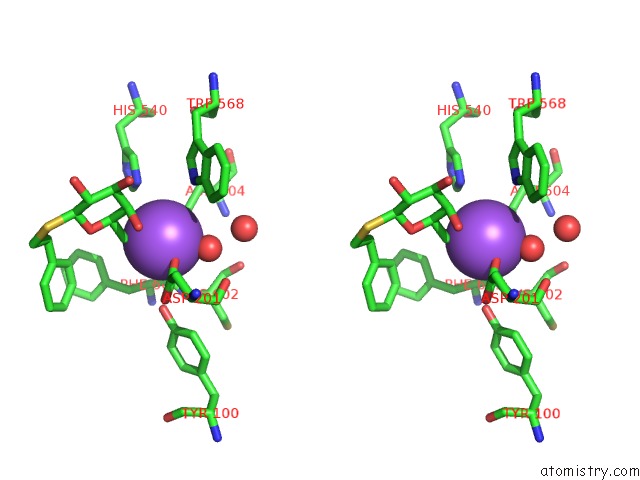
Sodium binding site 3 out of 8 in 5a1a
Go back to
Sodium binding site 3 out
of 8 in the 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with A Cell-Permeant Inhibitor
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 3 of 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with A Cell-Permeant Inhibitor within 5.0Å range:
|
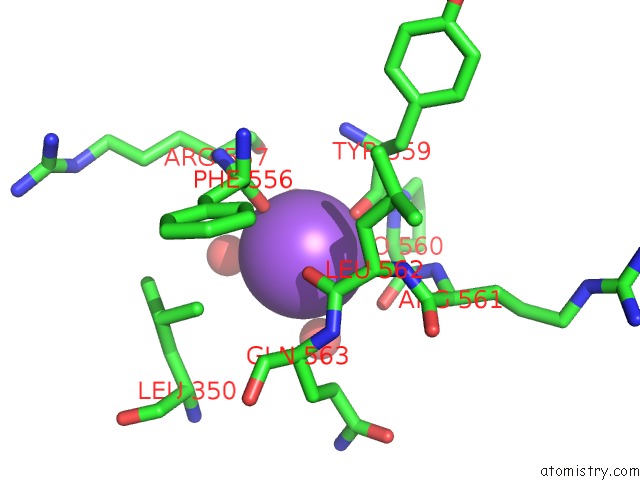
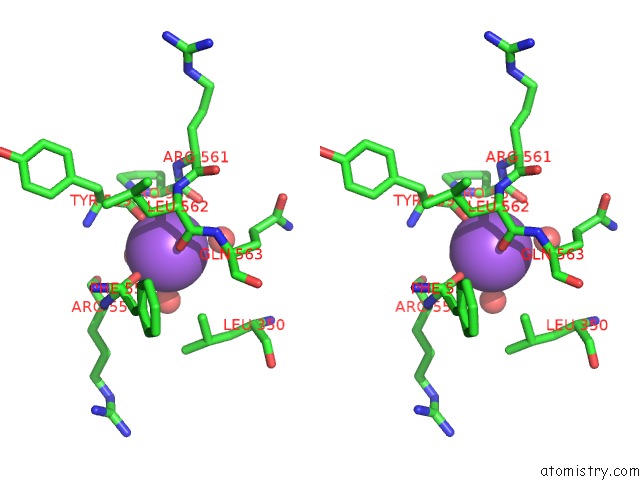
Sodium binding site 4 out of 8 in 5a1a
Go back to
Sodium binding site 4 out
of 8 in the 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with A Cell-Permeant Inhibitor
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 4 of 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with A Cell-Permeant Inhibitor within 5.0Å range:
|
Sodium binding site 5 out of 8 in 5a1a
Go back to
Sodium binding site 5 out
of 8 in the 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with A Cell-Permeant Inhibitor
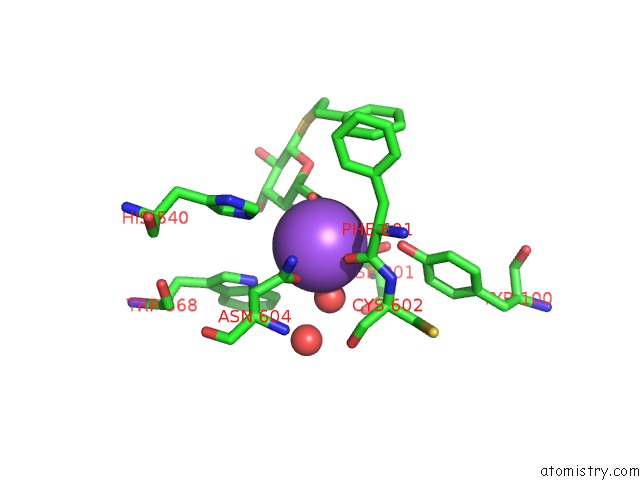
Mono view
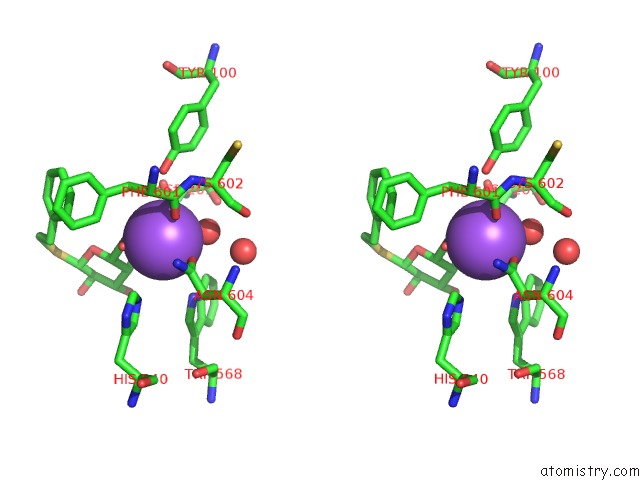
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 5 of 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with A Cell-Permeant Inhibitor within 5.0Å range:
|
Sodium binding site 6 out of 8 in 5a1a
Go back to
Sodium binding site 6 out
of 8 in the 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with A Cell-Permeant Inhibitor
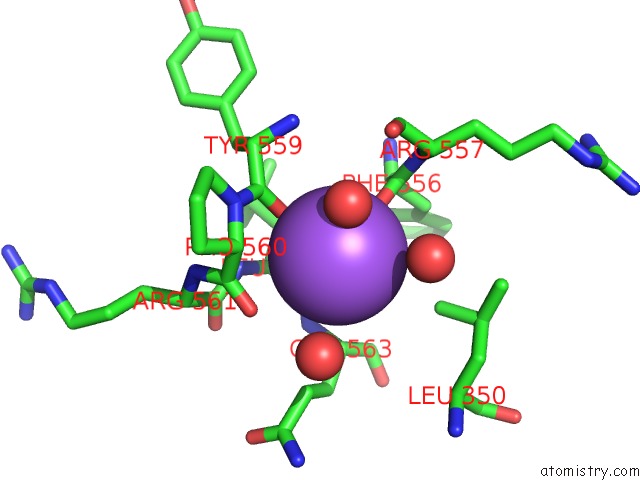
Mono view
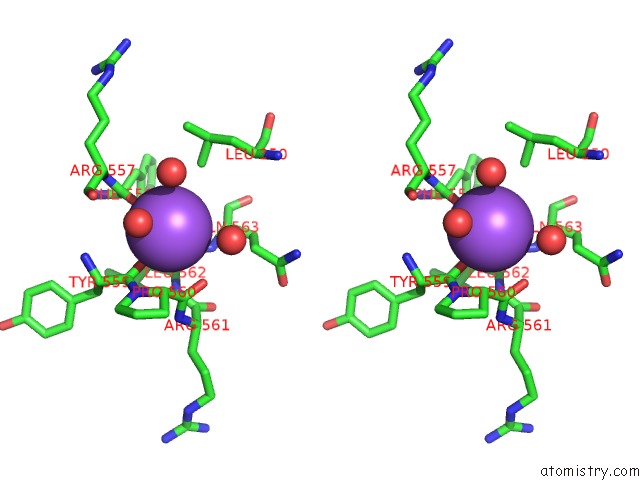
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 6 of 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with A Cell-Permeant Inhibitor within 5.0Å range:
|
Sodium binding site 7 out of 8 in 5a1a
Go back to
Sodium binding site 7 out
of 8 in the 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with A Cell-Permeant Inhibitor
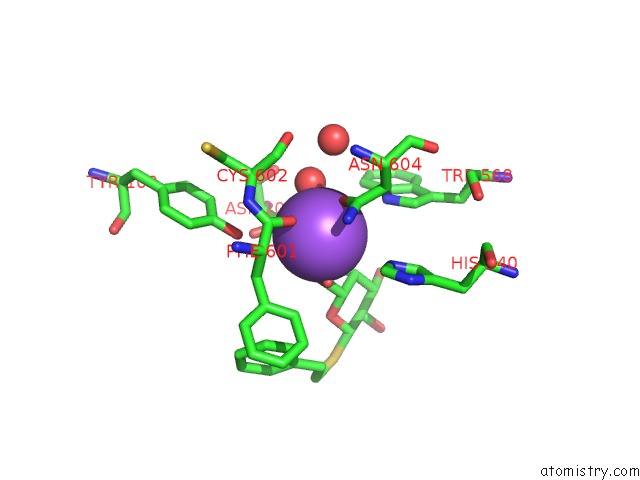
Mono view
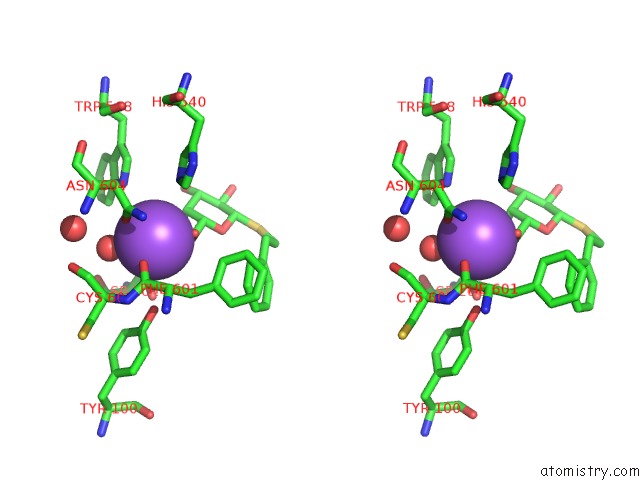
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 7 of 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with A Cell-Permeant Inhibitor within 5.0Å range:
|
Sodium binding site 8 out of 8 in 5a1a
Go back to
Sodium binding site 8 out
of 8 in the 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with A Cell-Permeant Inhibitor
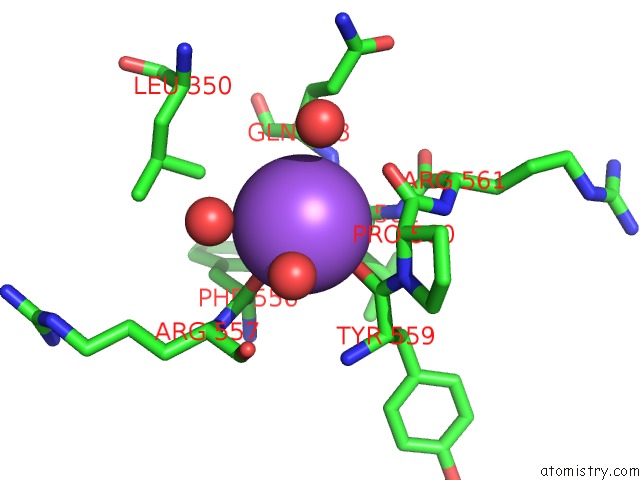
Mono view
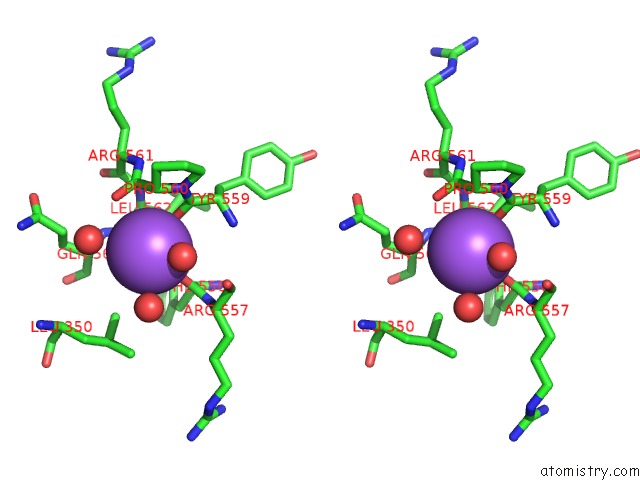
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 8 of 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with A Cell-Permeant Inhibitor within 5.0Å range:
|
Reference:
A.Bartesaghi,
A.Merk,
S.Banerjee,
D.Matthies,
X.Wu,
J.Milne,
S.Subramaniam.
2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with A Cell-Permeant Inhibitor Science 2015.
ISSN: ESSN 1095-9203
PubMed: 25953817
DOI: 10.1126/SCIENCE.AAB1576
Page generated: Mon Oct 7 19:48:21 2024
ISSN: ESSN 1095-9203
PubMed: 25953817
DOI: 10.1126/SCIENCE.AAB1576
Last articles
Cl in 5EQACl in 5ESG
Cl in 5ESH
Cl in 5ERK
Cl in 5ERJ
Cl in 5ERM
Cl in 5ER8
Cl in 5EPH
Cl in 5EQS
Cl in 5EQB