Sodium »
PDB 4yil-4z48 »
4ysa »
Sodium in PDB 4ysa: Completely Oxidized Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans
Protein crystallography data
The structure of Completely Oxidized Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans, PDB code: 4ysa
was solved by
Y.Fukuda,
K.M.Tse,
M.Suzuki,
K.Diederichs,
K.Hirata,
T.Nakane,
M.Sugahara,
E.Nango,
K.Tono,
Y.Joti,
T.Kameshima,
C.Song,
T.Hatsui,
M.Yabashi,
O.Nureki,
H.Matsumura,
T.Inoue,
S.Iwata,
E.Mizohata,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 34.75 / 1.43 |
| Space group | H 3 |
| Cell size a, b, c (Å), α, β, γ (°) | 116.197, 116.197, 85.553, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 13.7 / 14.9 |
Other elements in 4ysa:
The structure of Completely Oxidized Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans also contains other interesting chemical elements:
| Copper | (Cu) | 8 atoms |
Sodium Binding Sites:
The binding sites of Sodium atom in the Completely Oxidized Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans
(pdb code 4ysa). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Completely Oxidized Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans, PDB code: 4ysa:
In total only one binding site of Sodium was determined in the Completely Oxidized Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans, PDB code: 4ysa:
Sodium binding site 1 out of 1 in 4ysa
Go back to
Sodium binding site 1 out
of 1 in the Completely Oxidized Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans
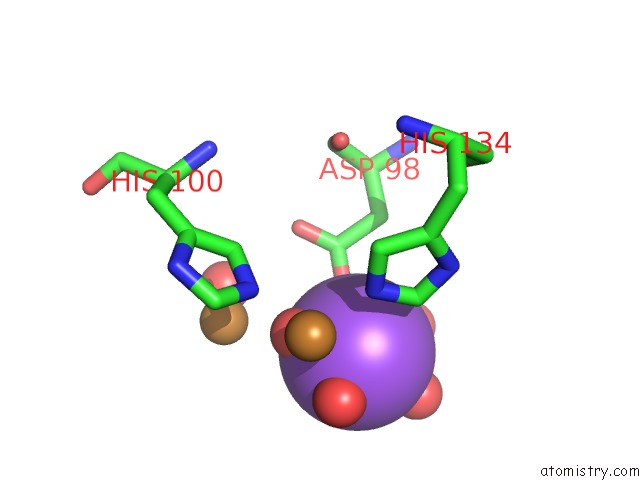
Mono view
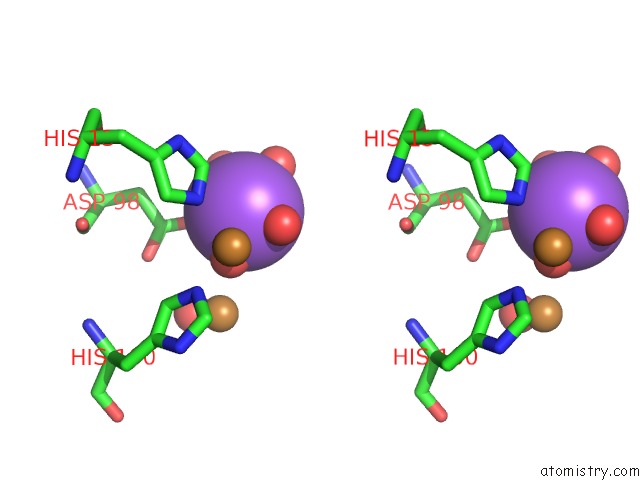
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Completely Oxidized Structure of Copper Nitrite Reductase From Geobacillus Thermodenitrificans within 5.0Å range:
|
Reference:
Y.Fukuda,
K.M.Tse,
M.Suzuki,
K.Diederichs,
K.Hirata,
T.Nakane,
M.Sugahara,
E.Nango,
K.Tono,
Y.Joti,
T.Kameshima,
C.Song,
T.Hatsui,
M.Yabashi,
O.Nureki,
H.Matsumura,
T.Inoue,
S.Iwata,
E.Mizohata.
Redox-Coupled Structural Changes in Nitrite Reductase Revealed By Serial Femtosecond and Microfocus Crystallography J.Biochem. V. 159 527 2016.
ISSN: ISSN 0021-924X
PubMed: 26769972
DOI: 10.1093/JB/MVV133
Page generated: Mon Oct 7 19:34:21 2024
ISSN: ISSN 0021-924X
PubMed: 26769972
DOI: 10.1093/JB/MVV133
Last articles
Ca in 5MTWCa in 5MQP
Ca in 5MU1
Ca in 5MST
Ca in 5MS4
Ca in 5MS9
Ca in 5MQR
Ca in 5MQS
Ca in 5MS3
Ca in 5MQM