Sodium »
PDB 4tmw-4u9k »
4tro »
Sodium in PDB 4tro: Structure of the Enoyl-Acp Reductase of Mycobacterium Tuberculosis Inha, Inhibited with the Active Metabolite of Isoniazid
Enzymatic activity of Structure of the Enoyl-Acp Reductase of Mycobacterium Tuberculosis Inha, Inhibited with the Active Metabolite of Isoniazid
All present enzymatic activity of Structure of the Enoyl-Acp Reductase of Mycobacterium Tuberculosis Inha, Inhibited with the Active Metabolite of Isoniazid:
1.3.1.9;
1.3.1.9;
Protein crystallography data
The structure of Structure of the Enoyl-Acp Reductase of Mycobacterium Tuberculosis Inha, Inhibited with the Active Metabolite of Isoniazid, PDB code: 4tro
was solved by
A.Chollet,
S.Julien,
L.Mourey,
L.Maveyraud,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 40.00 / 1.40 |
| Space group | P 62 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 97.434, 97.434, 139.752, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 12.1 / 15.1 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Structure of the Enoyl-Acp Reductase of Mycobacterium Tuberculosis Inha, Inhibited with the Active Metabolite of Isoniazid
(pdb code 4tro). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 2 binding sites of Sodium where determined in the Structure of the Enoyl-Acp Reductase of Mycobacterium Tuberculosis Inha, Inhibited with the Active Metabolite of Isoniazid, PDB code: 4tro:
Jump to Sodium binding site number: 1; 2;
In total 2 binding sites of Sodium where determined in the Structure of the Enoyl-Acp Reductase of Mycobacterium Tuberculosis Inha, Inhibited with the Active Metabolite of Isoniazid, PDB code: 4tro:
Jump to Sodium binding site number: 1; 2;
Sodium binding site 1 out of 2 in 4tro
Go back to
Sodium binding site 1 out
of 2 in the Structure of the Enoyl-Acp Reductase of Mycobacterium Tuberculosis Inha, Inhibited with the Active Metabolite of Isoniazid
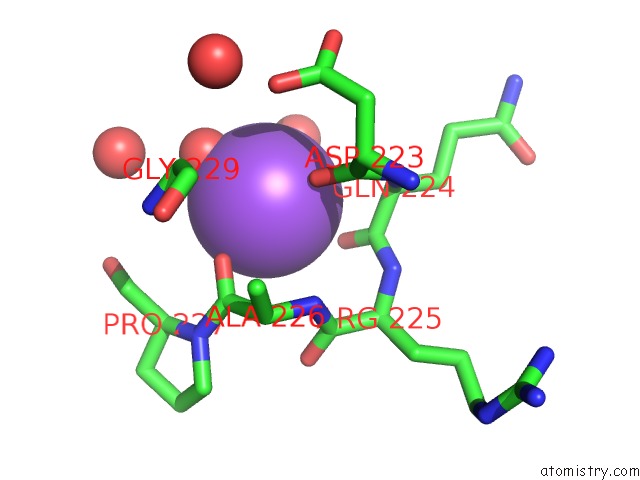
Mono view
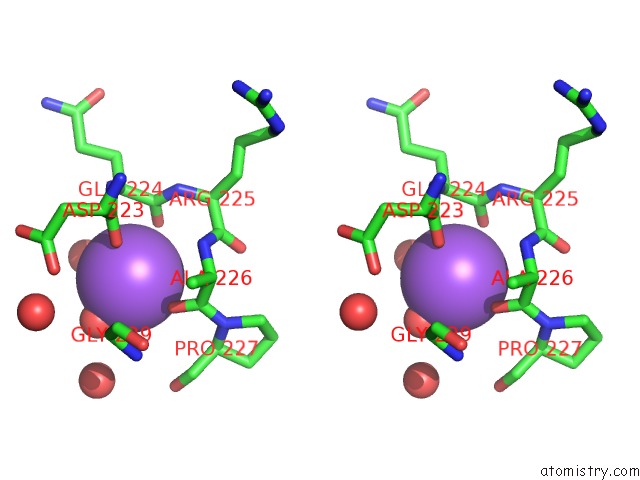
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Structure of the Enoyl-Acp Reductase of Mycobacterium Tuberculosis Inha, Inhibited with the Active Metabolite of Isoniazid within 5.0Å range:
|
Sodium binding site 2 out of 2 in 4tro
Go back to
Sodium binding site 2 out
of 2 in the Structure of the Enoyl-Acp Reductase of Mycobacterium Tuberculosis Inha, Inhibited with the Active Metabolite of Isoniazid

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Structure of the Enoyl-Acp Reductase of Mycobacterium Tuberculosis Inha, Inhibited with the Active Metabolite of Isoniazid within 5.0Å range:
|
Reference:
A.Chollet,
L.Mourey,
C.Lherbet,
A.Delbot,
S.Julien,
M.Baltas,
J.Bernadou,
G.Pratviel,
L.Maveyraud,
V.Bernardes-Genisson.
Crystal Structure of the Enoyl-Acp Reductase of Mycobacterium Tuberculosis (Inha) in the Apo-Form and in Complex with the Active Metabolite of Isoniazid Pre-Formed By A Biomimetic Approach. J.Struct.Biol. 2015.
ISSN: ESSN 1095-8657
PubMed: 25891098
DOI: 10.1016/J.JSB.2015.04.008
Page generated: Sun Aug 17 21:48:22 2025
ISSN: ESSN 1095-8657
PubMed: 25891098
DOI: 10.1016/J.JSB.2015.04.008
Last articles
Ni in 4MSNNi in 4N00
Ni in 4MTQ
Ni in 4MSH
Ni in 4MTT
Ni in 4MTS
Ni in 4MSC
Ni in 4MSE
Ni in 4MSA
Ni in 4MS0