Sodium »
PDB 4n01-4nji »
4n49 »
Sodium in PDB 4n49: Cap-Specific Mrna (Nucleoside-2'-O-)-Methyltransferase 1 Protein in Complex with M7GPPPG and Sam
Enzymatic activity of Cap-Specific Mrna (Nucleoside-2'-O-)-Methyltransferase 1 Protein in Complex with M7GPPPG and Sam
All present enzymatic activity of Cap-Specific Mrna (Nucleoside-2'-O-)-Methyltransferase 1 Protein in Complex with M7GPPPG and Sam:
2.1.1.57;
2.1.1.57;
Protein crystallography data
The structure of Cap-Specific Mrna (Nucleoside-2'-O-)-Methyltransferase 1 Protein in Complex with M7GPPPG and Sam, PDB code: 4n49
was solved by
M.Smietanski,
M.Werener,
E.Purta,
K.H.Kaminska,
J.Stepinski,
E.Darzynkiewicz,
M.Nowotny,
J.M.Bujnicki,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 46.80 / 1.90 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 50.834, 87.619, 57.310, 90.00, 112.89, 90.00 |
| R / Rfree (%) | 15.4 / 20.3 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Cap-Specific Mrna (Nucleoside-2'-O-)-Methyltransferase 1 Protein in Complex with M7GPPPG and Sam
(pdb code 4n49). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Cap-Specific Mrna (Nucleoside-2'-O-)-Methyltransferase 1 Protein in Complex with M7GPPPG and Sam, PDB code: 4n49:
In total only one binding site of Sodium was determined in the Cap-Specific Mrna (Nucleoside-2'-O-)-Methyltransferase 1 Protein in Complex with M7GPPPG and Sam, PDB code: 4n49:
Sodium binding site 1 out of 1 in 4n49
Go back to
Sodium binding site 1 out
of 1 in the Cap-Specific Mrna (Nucleoside-2'-O-)-Methyltransferase 1 Protein in Complex with M7GPPPG and Sam
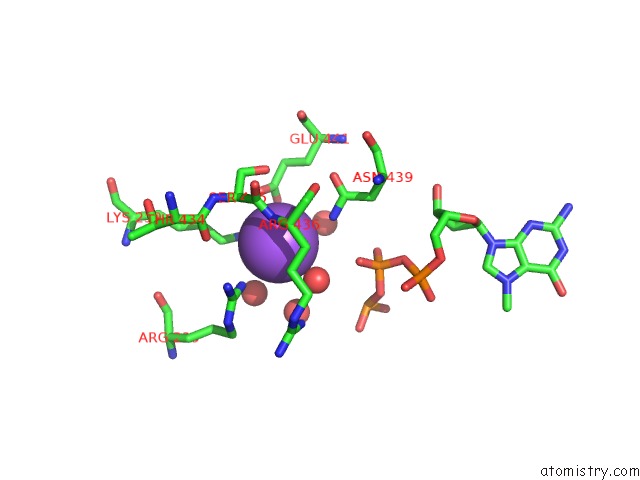
Mono view
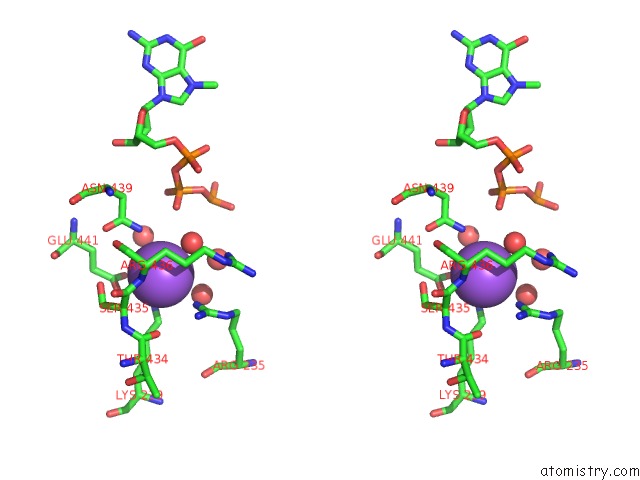
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Cap-Specific Mrna (Nucleoside-2'-O-)-Methyltransferase 1 Protein in Complex with M7GPPPG and Sam within 5.0Å range:
|
Reference:
M.Smietanski,
M.Werner,
E.Purta,
K.H.Kaminska,
J.Stepinski,
E.Darzynkiewicz,
M.Nowotny,
J.M.Bujnicki.
Structural Analysis of Human 2'-O-Ribose Methyltransferases Involved in Mrna Cap Structure Formation. Nat Commun V. 5 3004 2014.
ISSN: ESSN 2041-1723
PubMed: 24402442
DOI: 10.1038/NCOMMS4004
Page generated: Sun Aug 17 20:44:14 2025
ISSN: ESSN 2041-1723
PubMed: 24402442
DOI: 10.1038/NCOMMS4004
Last articles
Na in 6I0CNa in 6HZF
Na in 6I03
Na in 6HZG
Na in 6HZD
Na in 6HZ1
Na in 6HZB
Na in 6HZC
Na in 6HZA
Na in 6HZ0