Sodium »
PDB 4jvl-4khs »
4k6a »
Sodium in PDB 4k6a: Revised Crystal Structure of Apo-Form of Triosephosphate Isomerase (Tpia) From Escherichia Coli at 1.8 Angstrom Resolution.
Enzymatic activity of Revised Crystal Structure of Apo-Form of Triosephosphate Isomerase (Tpia) From Escherichia Coli at 1.8 Angstrom Resolution.
All present enzymatic activity of Revised Crystal Structure of Apo-Form of Triosephosphate Isomerase (Tpia) From Escherichia Coli at 1.8 Angstrom Resolution.:
5.3.1.1;
5.3.1.1;
Protein crystallography data
The structure of Revised Crystal Structure of Apo-Form of Triosephosphate Isomerase (Tpia) From Escherichia Coli at 1.8 Angstrom Resolution., PDB code: 4k6a
was solved by
G.Minasov,
M.Kuhn,
A.Halavaty,
L.Shuvalova,
I.Dubrovska,
J.Winsor,
S.Grimshaw,
W.F.Anderson,
Center For Structural Genomics Of Infectiousdiseases (Csgid),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 29.06 / 1.80 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 46.066, 67.488, 149.769, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 14.9 / 18.5 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Revised Crystal Structure of Apo-Form of Triosephosphate Isomerase (Tpia) From Escherichia Coli at 1.8 Angstrom Resolution.
(pdb code 4k6a). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Revised Crystal Structure of Apo-Form of Triosephosphate Isomerase (Tpia) From Escherichia Coli at 1.8 Angstrom Resolution., PDB code: 4k6a:
In total only one binding site of Sodium was determined in the Revised Crystal Structure of Apo-Form of Triosephosphate Isomerase (Tpia) From Escherichia Coli at 1.8 Angstrom Resolution., PDB code: 4k6a:
Sodium binding site 1 out of 1 in 4k6a
Go back to
Sodium binding site 1 out
of 1 in the Revised Crystal Structure of Apo-Form of Triosephosphate Isomerase (Tpia) From Escherichia Coli at 1.8 Angstrom Resolution.
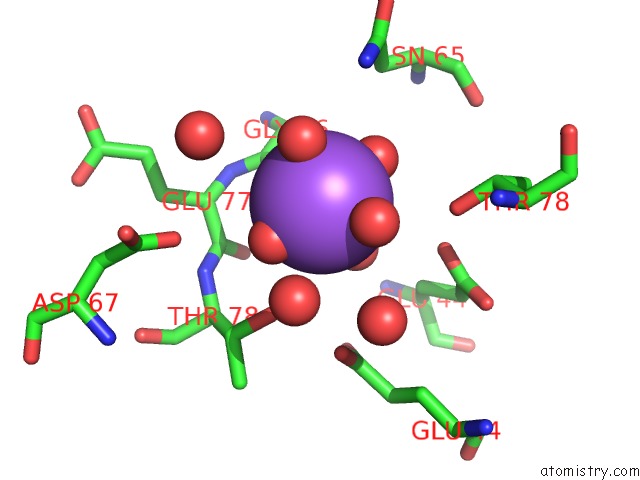
Mono view
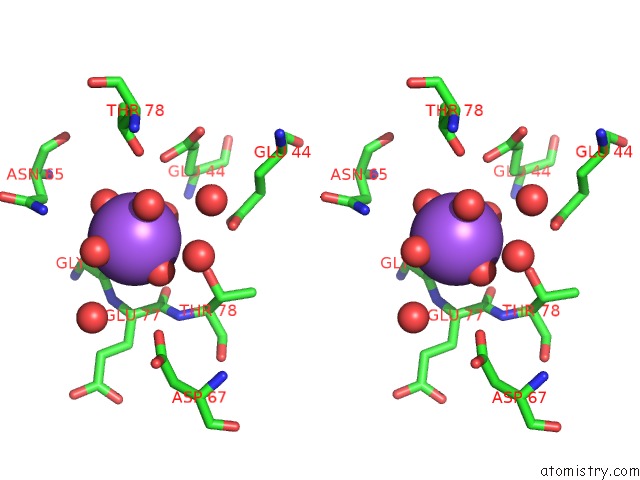
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Revised Crystal Structure of Apo-Form of Triosephosphate Isomerase (Tpia) From Escherichia Coli at 1.8 Angstrom Resolution. within 5.0Å range:
|
Reference:
M.L.Kuhn,
B.Zemaitaitis,
L.I.Hu,
A.Sahu,
D.Sorensen,
G.Minasov,
B.P.Lima,
M.Scholle,
M.Mrksich,
W.F.Anderson,
B.W.Gibson,
B.Schilling,
A.J.Wolfe.
Structural, Kinetic and Proteomic Characterization of Acetyl Phosphate-Dependent Bacterial Protein Acetylation. Plos One V. 9 94816 2014.
ISSN: ESSN 1932-6203
PubMed: 24756028
DOI: 10.1371/JOURNAL.PONE.0094816
Page generated: Mon Oct 7 16:24:43 2024
ISSN: ESSN 1932-6203
PubMed: 24756028
DOI: 10.1371/JOURNAL.PONE.0094816
Last articles
Ca in 5NRKCa in 5NUR
Ca in 5NSF
Ca in 5NRM
Ca in 5NUC
Ca in 5NRT
Ca in 5NSA
Ca in 5NPF
Ca in 5NQ2
Ca in 5N97