Sodium »
PDB 4blw-4c44 »
4c1p »
Sodium in PDB 4c1p: Geobacillus Thermoglucosidasius Gh Family 52 Xylosidase
Enzymatic activity of Geobacillus Thermoglucosidasius Gh Family 52 Xylosidase
All present enzymatic activity of Geobacillus Thermoglucosidasius Gh Family 52 Xylosidase:
3.2.1.37;
3.2.1.37;
Protein crystallography data
The structure of Geobacillus Thermoglucosidasius Gh Family 52 Xylosidase, PDB code: 4c1p
was solved by
G.Espina,
K.Eley,
T.R.Schneider,
S.J.Crennell,
M.J.Danson,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 27.311 / 2.63 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 73.369, 105.335, 194.834, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.23 / 23.8 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Geobacillus Thermoglucosidasius Gh Family 52 Xylosidase
(pdb code 4c1p). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Geobacillus Thermoglucosidasius Gh Family 52 Xylosidase, PDB code: 4c1p:
In total only one binding site of Sodium was determined in the Geobacillus Thermoglucosidasius Gh Family 52 Xylosidase, PDB code: 4c1p:
Sodium binding site 1 out of 1 in 4c1p
Go back to
Sodium binding site 1 out
of 1 in the Geobacillus Thermoglucosidasius Gh Family 52 Xylosidase
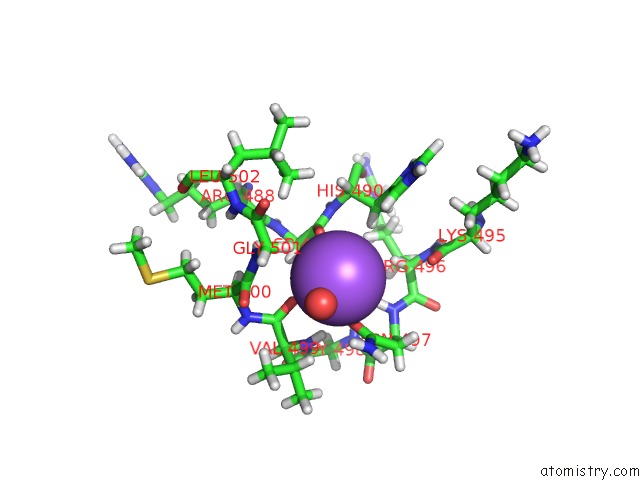
Mono view
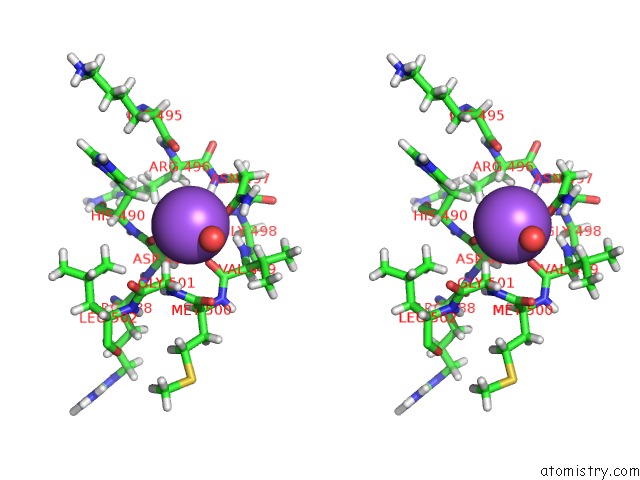
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Geobacillus Thermoglucosidasius Gh Family 52 Xylosidase within 5.0Å range:
|
Reference:
G.Espina,
K.Eley,
G.Pompidor,
T.R.Schneider,
S.J.Crennell,
M.J.Danson.
A Novel Beta-Xylosidase Structure From Geobacillus Thermoglucosidasius: the First Crystal Structure of A Glycoside Hydrolase Family GH52 Enzyme Reveals Unpredicted Similarity to Other Glycoside Hydrolase Folds Acta Crystallogr.,Sect.D V. 70 1366 2014.
ISSN: ISSN 0907-4449
PubMed: 24816105
DOI: 10.1107/S1399004714002788
Page generated: Sun Aug 17 18:42:39 2025
ISSN: ISSN 0907-4449
PubMed: 24816105
DOI: 10.1107/S1399004714002788
Last articles
Na in 4OMHNa in 4OKI
Na in 4OMG
Na in 4OLF
Na in 4OLN
Na in 4OHC
Na in 4OGL
Na in 4OJ1
Na in 4OFI
Na in 4OIC