Sodium »
PDB 4b1a-4bkr »
4bdn »
Sodium in PDB 4bdn: Crystal Structure of the GLUK2 K531A-T779G Lbd Dimer in Complex with Glutamate
Protein crystallography data
The structure of Crystal Structure of the GLUK2 K531A-T779G Lbd Dimer in Complex with Glutamate, PDB code: 4bdn
was solved by
N.Nayeem,
O.Mayans,
T.Green,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 32.561 / 2.50 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 85.729, 100.133, 126.199, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19.75 / 24.79 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Crystal Structure of the GLUK2 K531A-T779G Lbd Dimer in Complex with Glutamate
(pdb code 4bdn). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 4 binding sites of Sodium where determined in the Crystal Structure of the GLUK2 K531A-T779G Lbd Dimer in Complex with Glutamate, PDB code: 4bdn:
Jump to Sodium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Sodium where determined in the Crystal Structure of the GLUK2 K531A-T779G Lbd Dimer in Complex with Glutamate, PDB code: 4bdn:
Jump to Sodium binding site number: 1; 2; 3; 4;
Sodium binding site 1 out of 4 in 4bdn
Go back to
Sodium binding site 1 out
of 4 in the Crystal Structure of the GLUK2 K531A-T779G Lbd Dimer in Complex with Glutamate
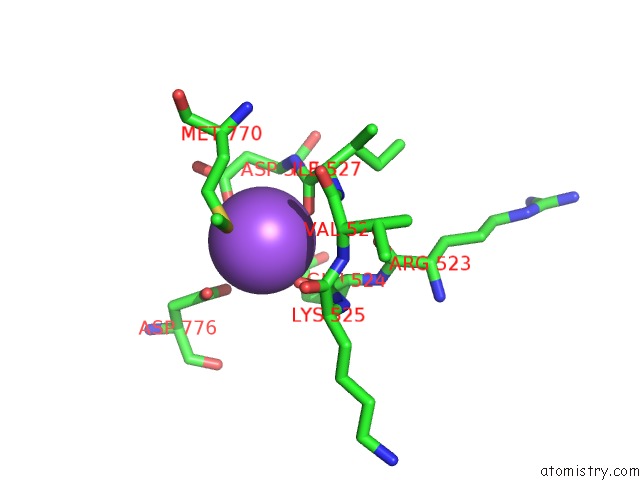
Mono view
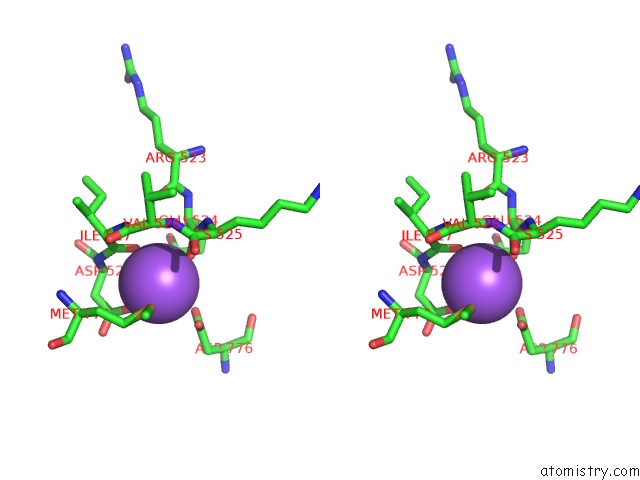
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Crystal Structure of the GLUK2 K531A-T779G Lbd Dimer in Complex with Glutamate within 5.0Å range:
|
Sodium binding site 2 out of 4 in 4bdn
Go back to
Sodium binding site 2 out
of 4 in the Crystal Structure of the GLUK2 K531A-T779G Lbd Dimer in Complex with Glutamate
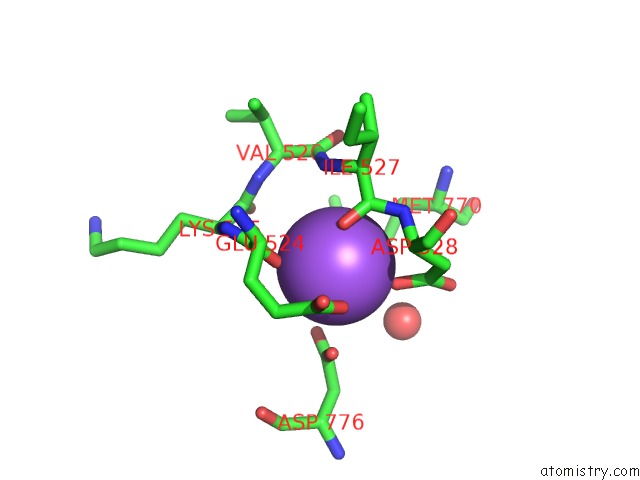
Mono view
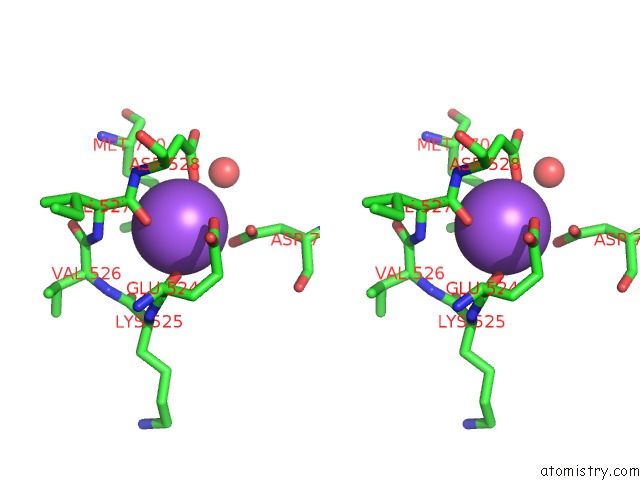
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Crystal Structure of the GLUK2 K531A-T779G Lbd Dimer in Complex with Glutamate within 5.0Å range:
|
Sodium binding site 3 out of 4 in 4bdn
Go back to
Sodium binding site 3 out
of 4 in the Crystal Structure of the GLUK2 K531A-T779G Lbd Dimer in Complex with Glutamate
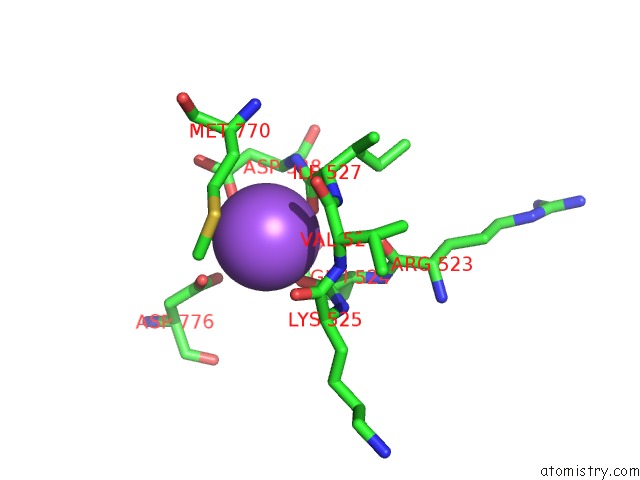
Mono view
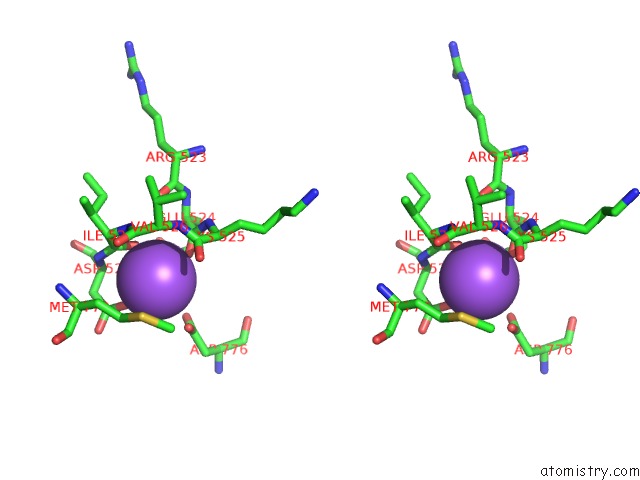
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 3 of Crystal Structure of the GLUK2 K531A-T779G Lbd Dimer in Complex with Glutamate within 5.0Å range:
|
Sodium binding site 4 out of 4 in 4bdn
Go back to
Sodium binding site 4 out
of 4 in the Crystal Structure of the GLUK2 K531A-T779G Lbd Dimer in Complex with Glutamate
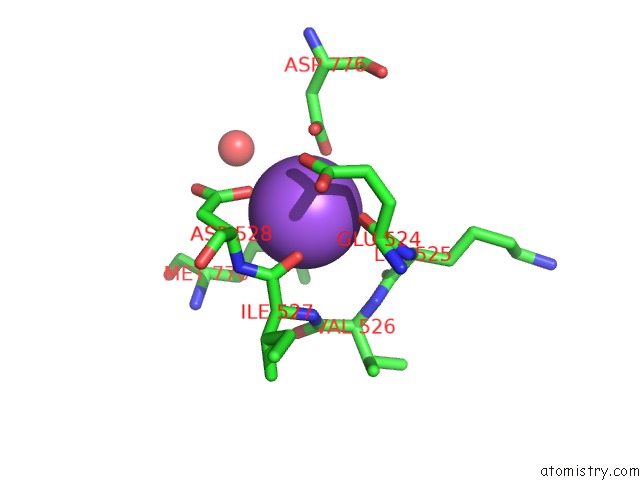
Mono view
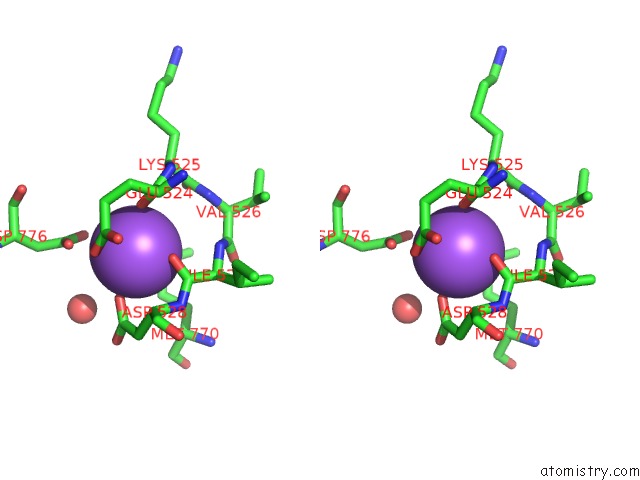
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 4 of Crystal Structure of the GLUK2 K531A-T779G Lbd Dimer in Complex with Glutamate within 5.0Å range:
|
Reference:
N.Nayeem,
O.Mayans,
T.Green.
Correlating Efficacy and Desensitization with GLUK2 Ligand- Binding Domain Movements. Open Biol. V. 3 0051 2013.
ISSN: ISSN 2046-2441
PubMed: 23720540
DOI: 10.1098/RSOB.130051
Page generated: Mon Oct 7 14:29:47 2024
ISSN: ISSN 2046-2441
PubMed: 23720540
DOI: 10.1098/RSOB.130051
Last articles
Cl in 7V91Cl in 7VBX
Cl in 7VB8
Cl in 7VBV
Cl in 7V8K
Cl in 7VBU
Cl in 7V73
Cl in 7V68
Cl in 7V57
Cl in 7V46