Sodium »
PDB 3t0a-3tjl »
3tf9 »
Sodium in PDB 3tf9: Crystal Structure of An H-Nox Protein From Nostoc Sp. Pcc 7120 Under 1 Atm of Xenon
Protein crystallography data
The structure of Crystal Structure of An H-Nox Protein From Nostoc Sp. Pcc 7120 Under 1 Atm of Xenon, PDB code: 3tf9
was solved by
M.B.Winter,
M.A.Herzik Jr.,
J.Kuriyan,
M.A.Marletta,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 43.98 / 2.59 |
| Space group | P 21 3 |
| Cell size a, b, c (Å), α, β, γ (°) | 124.404, 124.404, 124.404, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17 / 20.5 |
Other elements in 3tf9:
The structure of Crystal Structure of An H-Nox Protein From Nostoc Sp. Pcc 7120 Under 1 Atm of Xenon also contains other interesting chemical elements:
| Xenon | (Xe) | 2 atoms |
| Iron | (Fe) | 2 atoms |
Sodium Binding Sites:
The binding sites of Sodium atom in the Crystal Structure of An H-Nox Protein From Nostoc Sp. Pcc 7120 Under 1 Atm of Xenon
(pdb code 3tf9). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 2 binding sites of Sodium where determined in the Crystal Structure of An H-Nox Protein From Nostoc Sp. Pcc 7120 Under 1 Atm of Xenon, PDB code: 3tf9:
Jump to Sodium binding site number: 1; 2;
In total 2 binding sites of Sodium where determined in the Crystal Structure of An H-Nox Protein From Nostoc Sp. Pcc 7120 Under 1 Atm of Xenon, PDB code: 3tf9:
Jump to Sodium binding site number: 1; 2;
Sodium binding site 1 out of 2 in 3tf9
Go back to
Sodium binding site 1 out
of 2 in the Crystal Structure of An H-Nox Protein From Nostoc Sp. Pcc 7120 Under 1 Atm of Xenon
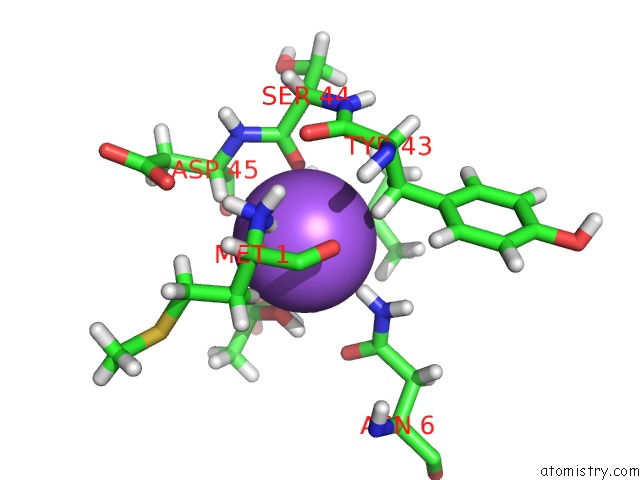
Mono view
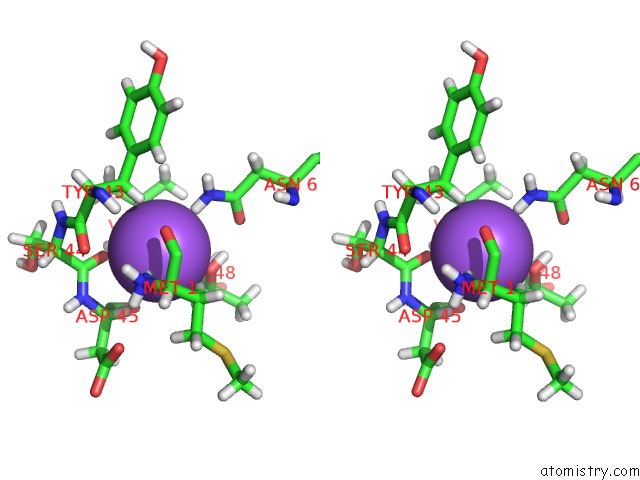
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Crystal Structure of An H-Nox Protein From Nostoc Sp. Pcc 7120 Under 1 Atm of Xenon within 5.0Å range:
|
Sodium binding site 2 out of 2 in 3tf9
Go back to
Sodium binding site 2 out
of 2 in the Crystal Structure of An H-Nox Protein From Nostoc Sp. Pcc 7120 Under 1 Atm of Xenon
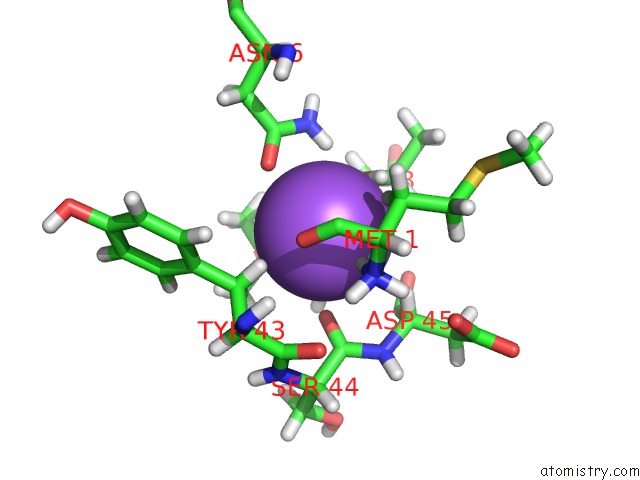
Mono view
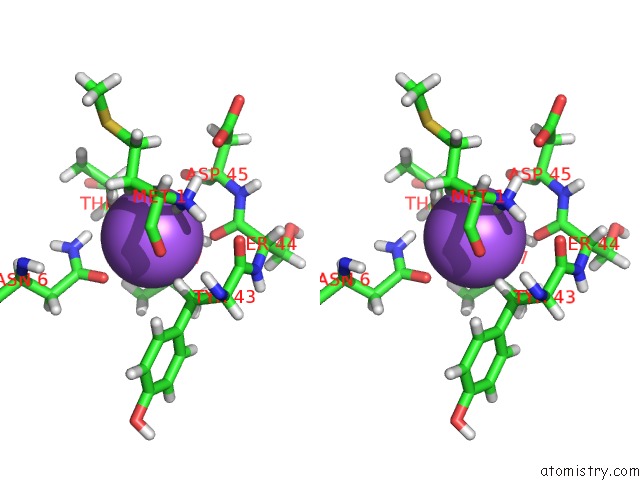
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Crystal Structure of An H-Nox Protein From Nostoc Sp. Pcc 7120 Under 1 Atm of Xenon within 5.0Å range:
|
Reference:
M.B.Winter,
M.A.Herzik,
J.Kuriyan,
M.A.Marletta.
Tunnels Modulate Ligand Flux in A Heme Nitric Oxide/Oxygen Binding (H-Nox) Domain. Proc.Natl.Acad.Sci.Usa V. 108 E881 2011.
ISSN: ISSN 0027-8424
PubMed: 21997213
DOI: 10.1073/PNAS.1114038108
Page generated: Mon Oct 7 13:16:28 2024
ISSN: ISSN 0027-8424
PubMed: 21997213
DOI: 10.1073/PNAS.1114038108
Last articles
Cl in 5TICCl in 5TI4
Cl in 5TIN
Cl in 5THP
Cl in 5TGZ
Cl in 5TGQ
Cl in 5TGN
Cl in 5TG3
Cl in 5TG1
Cl in 5TF6