Sodium »
PDB 3rnx-3si4 »
3s7x »
Sodium in PDB 3s7x: Unassembled Washington University Polyomavirus VP1 Pentamer R198K Mutant
Protein crystallography data
The structure of Unassembled Washington University Polyomavirus VP1 Pentamer R198K Mutant, PDB code: 3s7x
was solved by
U.Neu,
J.Wang,
T.Stehle,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 43.36 / 2.90 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 166.200, 166.200, 127.570, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 23.4 / 25.3 |
Other elements in 3s7x:
The structure of Unassembled Washington University Polyomavirus VP1 Pentamer R198K Mutant also contains other interesting chemical elements:
| Chlorine | (Cl) | 5 atoms |
Sodium Binding Sites:
The binding sites of Sodium atom in the Unassembled Washington University Polyomavirus VP1 Pentamer R198K Mutant
(pdb code 3s7x). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 5 binding sites of Sodium where determined in the Unassembled Washington University Polyomavirus VP1 Pentamer R198K Mutant, PDB code: 3s7x:
Jump to Sodium binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Sodium where determined in the Unassembled Washington University Polyomavirus VP1 Pentamer R198K Mutant, PDB code: 3s7x:
Jump to Sodium binding site number: 1; 2; 3; 4; 5;
Sodium binding site 1 out of 5 in 3s7x
Go back to
Sodium binding site 1 out
of 5 in the Unassembled Washington University Polyomavirus VP1 Pentamer R198K Mutant

Mono view
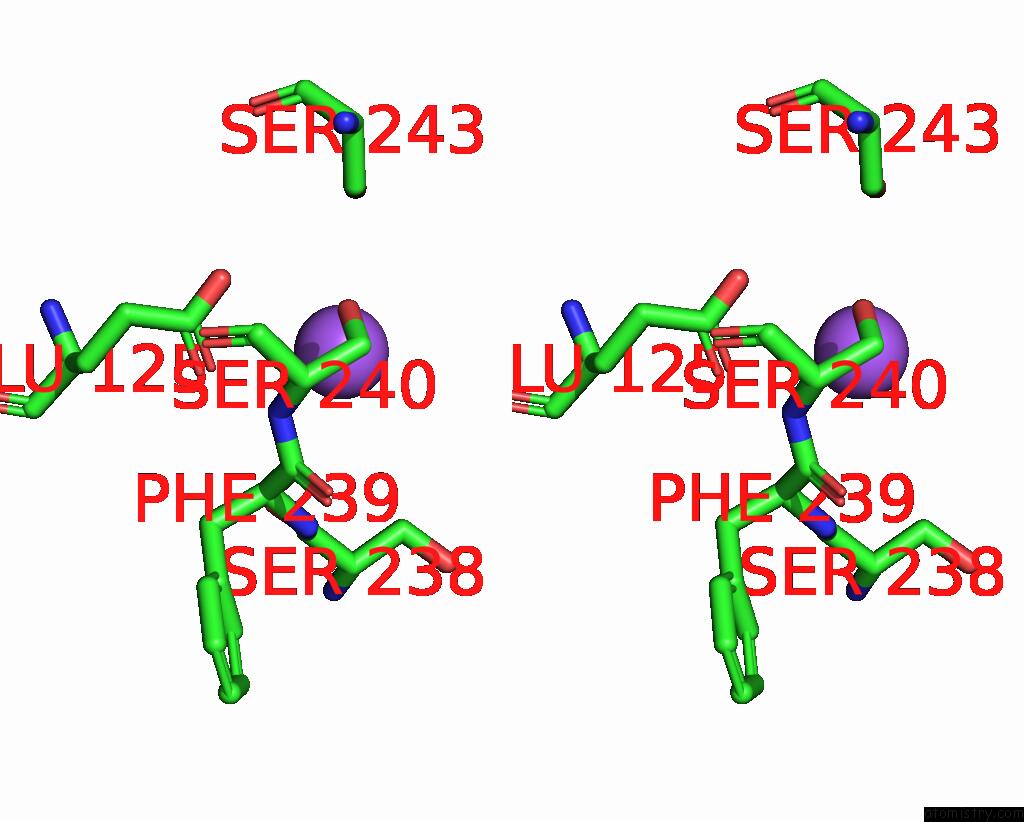
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Unassembled Washington University Polyomavirus VP1 Pentamer R198K Mutant within 5.0Å range:
|
Sodium binding site 2 out of 5 in 3s7x
Go back to
Sodium binding site 2 out
of 5 in the Unassembled Washington University Polyomavirus VP1 Pentamer R198K Mutant
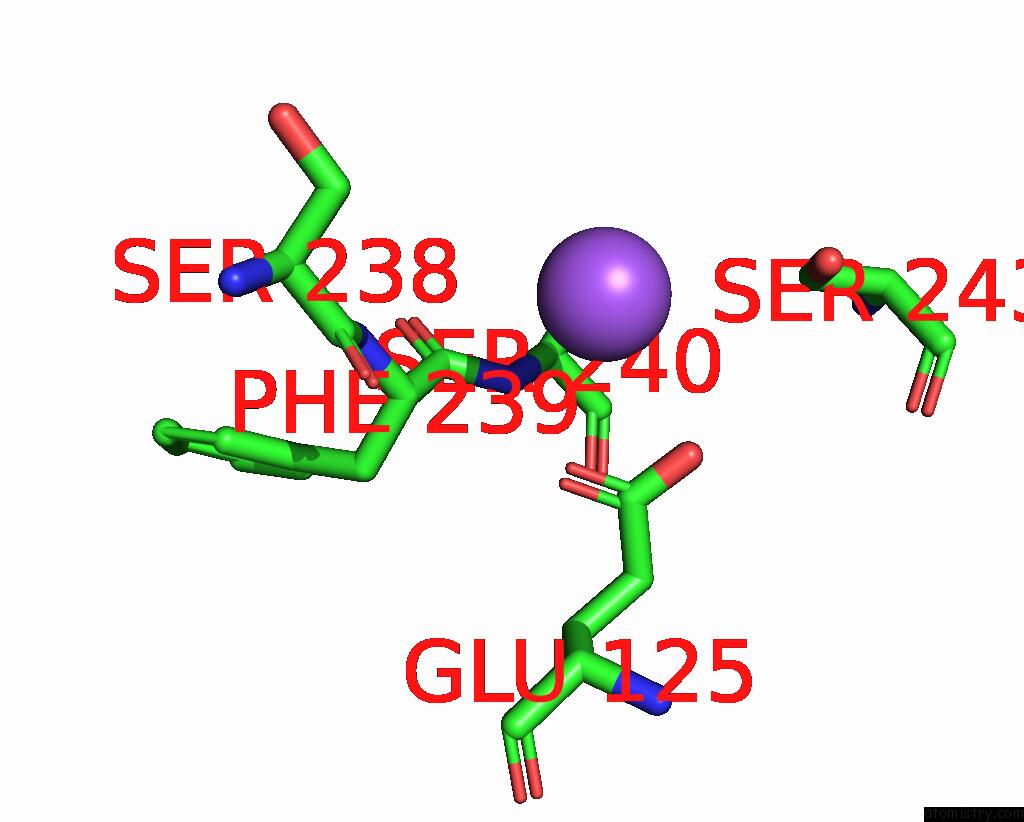
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Unassembled Washington University Polyomavirus VP1 Pentamer R198K Mutant within 5.0Å range:
|
Sodium binding site 3 out of 5 in 3s7x
Go back to
Sodium binding site 3 out
of 5 in the Unassembled Washington University Polyomavirus VP1 Pentamer R198K Mutant
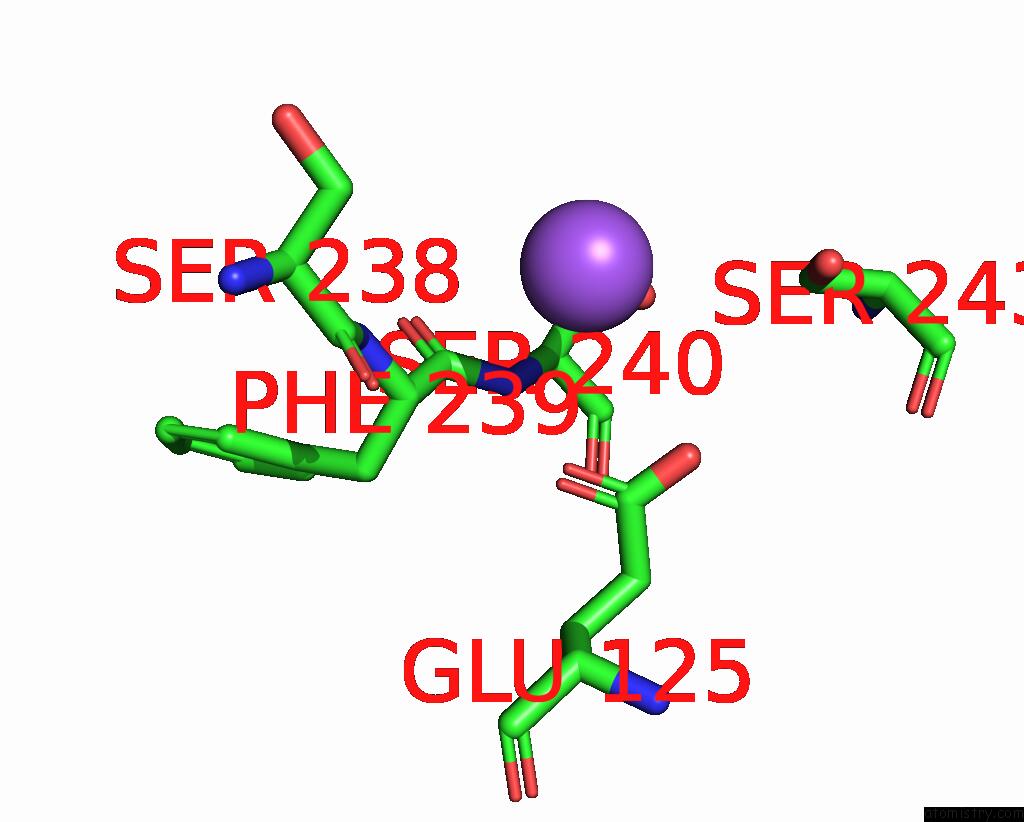
Mono view
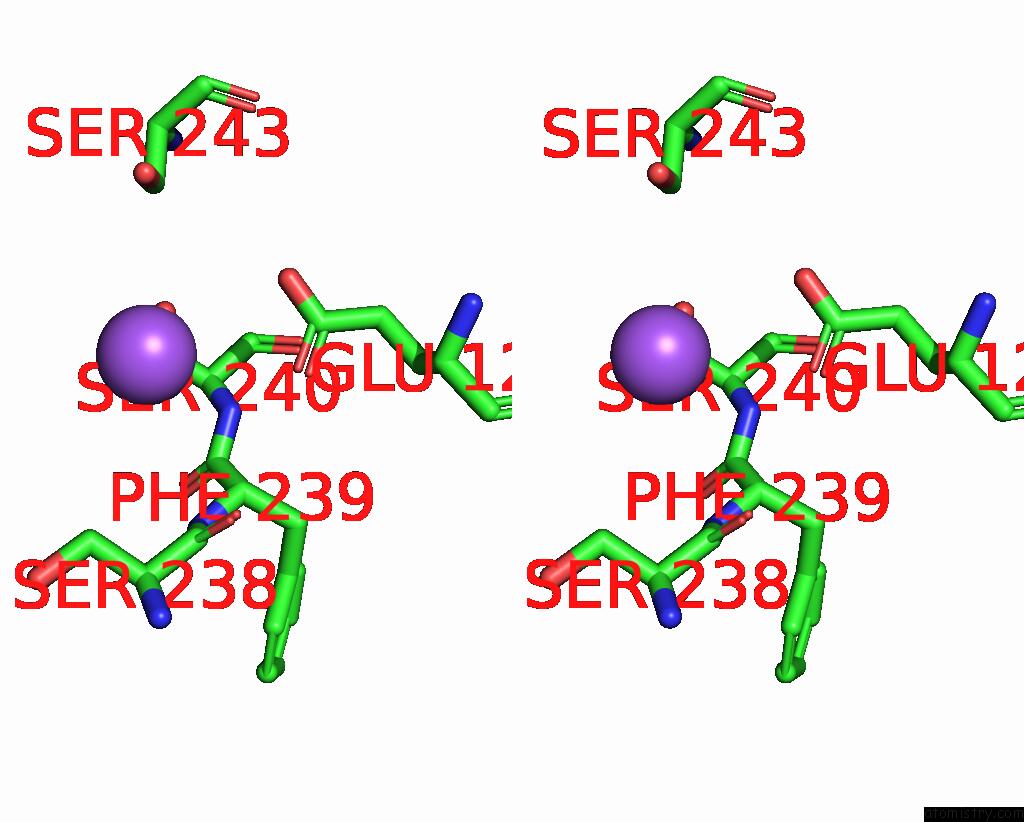
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 3 of Unassembled Washington University Polyomavirus VP1 Pentamer R198K Mutant within 5.0Å range:
|
Sodium binding site 4 out of 5 in 3s7x
Go back to
Sodium binding site 4 out
of 5 in the Unassembled Washington University Polyomavirus VP1 Pentamer R198K Mutant
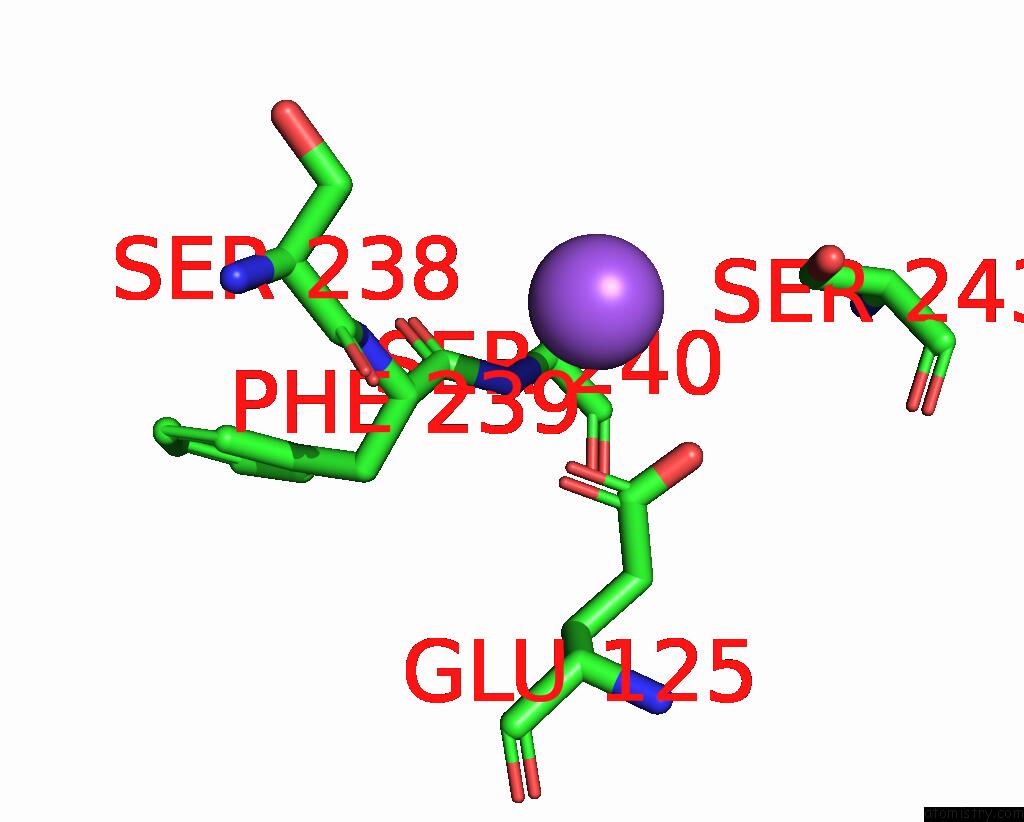
Mono view
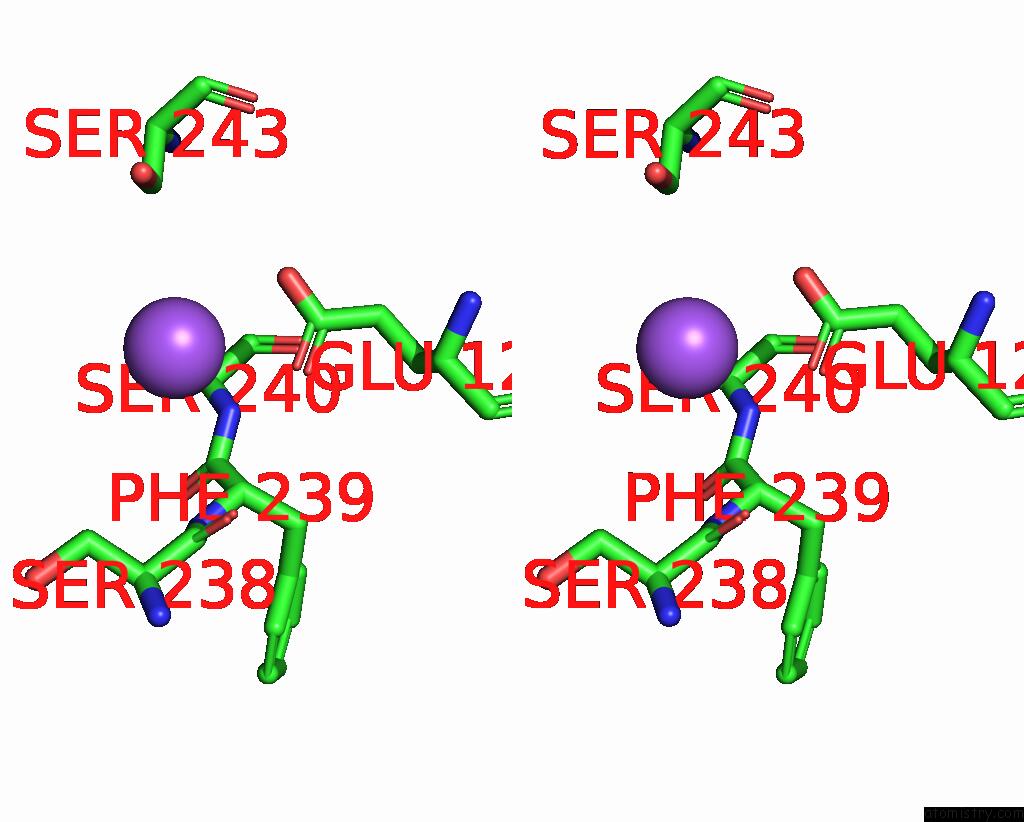
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 4 of Unassembled Washington University Polyomavirus VP1 Pentamer R198K Mutant within 5.0Å range:
|
Sodium binding site 5 out of 5 in 3s7x
Go back to
Sodium binding site 5 out
of 5 in the Unassembled Washington University Polyomavirus VP1 Pentamer R198K Mutant

Mono view
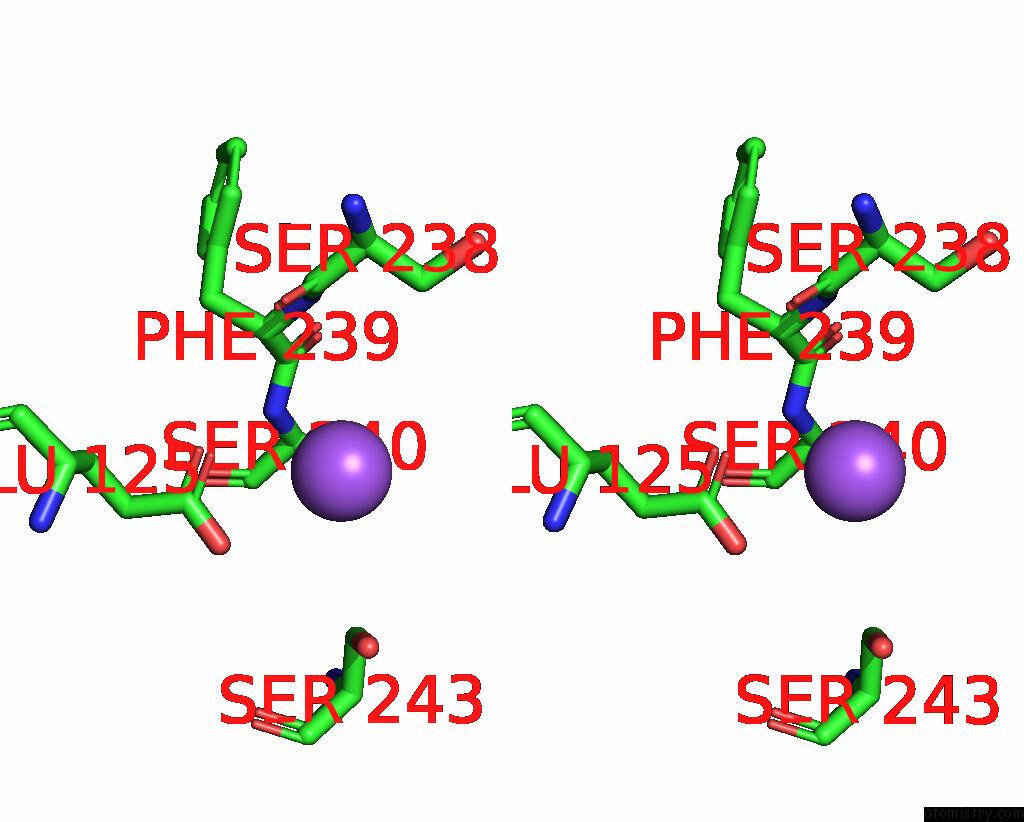
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 5 of Unassembled Washington University Polyomavirus VP1 Pentamer R198K Mutant within 5.0Å range:
|
Reference:
U.Neu,
J.Wang,
D.Macejak,
R.L.Garcea,
T.Stehle.
Structures of the Major Capsid Proteins of the Human Ki and Wu Polyomaviruses. J.Virol. V. 85 7384 2011.
ISSN: ISSN 0022-538X
PubMed: 21543504
DOI: 10.1128/JVI.00382-11
Page generated: Sun Aug 17 17:20:15 2025
ISSN: ISSN 0022-538X
PubMed: 21543504
DOI: 10.1128/JVI.00382-11
Last articles
Na in 7DLNNa in 7DFO
Na in 7DLH
Na in 7DJQ
Na in 7DJK
Na in 7DKM
Na in 7DJM
Na in 7DJL
Na in 7DJC
Na in 7DJJ