Sodium »
PDB 3iwf-3k93 »
3j7t »
Sodium in PDB 3j7t: Calcium Atpase Structure with Two Bound Calcium Ions Determined By Electron Crystallography of Thin 3D Crystals
Enzymatic activity of Calcium Atpase Structure with Two Bound Calcium Ions Determined By Electron Crystallography of Thin 3D Crystals
All present enzymatic activity of Calcium Atpase Structure with Two Bound Calcium Ions Determined By Electron Crystallography of Thin 3D Crystals:
3.6.3.8;
3.6.3.8;
Other elements in 3j7t:
The structure of Calcium Atpase Structure with Two Bound Calcium Ions Determined By Electron Crystallography of Thin 3D Crystals also contains other interesting chemical elements:
| Calcium | (Ca) | 2 atoms |
Sodium Binding Sites:
The binding sites of Sodium atom in the Calcium Atpase Structure with Two Bound Calcium Ions Determined By Electron Crystallography of Thin 3D Crystals
(pdb code 3j7t). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Calcium Atpase Structure with Two Bound Calcium Ions Determined By Electron Crystallography of Thin 3D Crystals, PDB code: 3j7t:
In total only one binding site of Sodium was determined in the Calcium Atpase Structure with Two Bound Calcium Ions Determined By Electron Crystallography of Thin 3D Crystals, PDB code: 3j7t:
Sodium binding site 1 out of 1 in 3j7t
Go back to
Sodium binding site 1 out
of 1 in the Calcium Atpase Structure with Two Bound Calcium Ions Determined By Electron Crystallography of Thin 3D Crystals
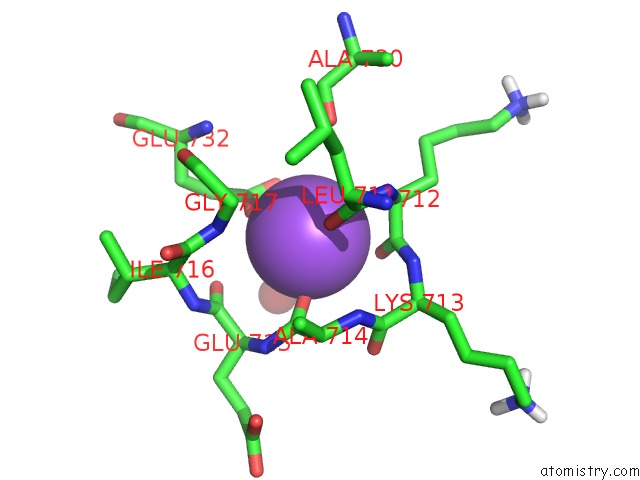
Mono view
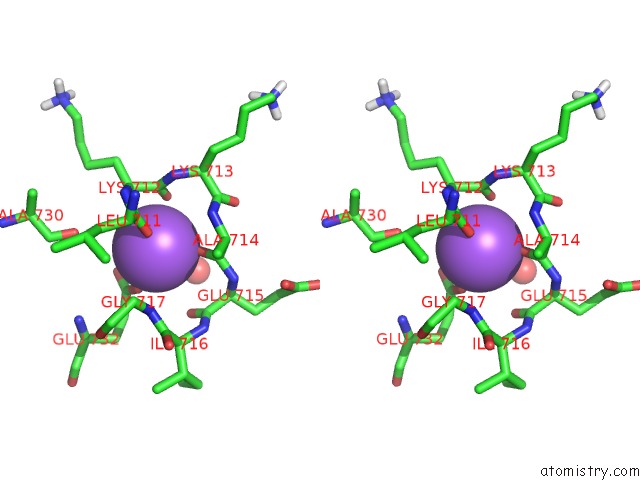
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Calcium Atpase Structure with Two Bound Calcium Ions Determined By Electron Crystallography of Thin 3D Crystals within 5.0Å range:
|
Reference:
K.Yonekura,
K.Kato,
M.Ogasawara,
M.Tomita,
C.Toyoshima.
Electron Crystallography of Ultra-Thin 3D Protein Crystals: Atomic Model with Charges Proc.Natl.Acad.Sci.Usa 2015.
ISSN: ESSN 1091-6490
Page generated: Mon Oct 7 11:04:13 2024
ISSN: ESSN 1091-6490
Last articles
Zn in 9JYWZn in 9IR4
Zn in 9IR3
Zn in 9GMX
Zn in 9GMW
Zn in 9JEJ
Zn in 9ERF
Zn in 9ERE
Zn in 9EGV
Zn in 9EGW