Sodium »
PDB 3cq8-3dfh »
3cqx »
Sodium in PDB 3cqx: Chaperone Complex
Protein crystallography data
The structure of Chaperone Complex, PDB code: 3cqx
was solved by
Z.Xu,
J.C.Nix,
S.Misra,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 43.13 / 2.30 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 52.846, 105.808, 210.683, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 22.2 / 25.6 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Chaperone Complex
(pdb code 3cqx). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 4 binding sites of Sodium where determined in the Chaperone Complex, PDB code: 3cqx:
Jump to Sodium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Sodium where determined in the Chaperone Complex, PDB code: 3cqx:
Jump to Sodium binding site number: 1; 2; 3; 4;
Sodium binding site 1 out of 4 in 3cqx
Go back to
Sodium binding site 1 out
of 4 in the Chaperone Complex
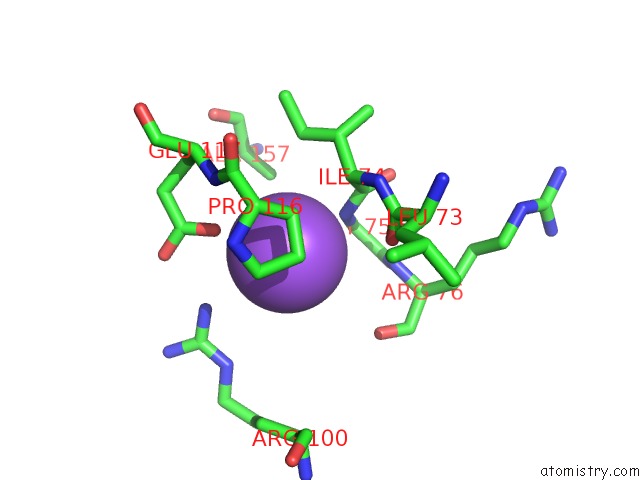
Mono view
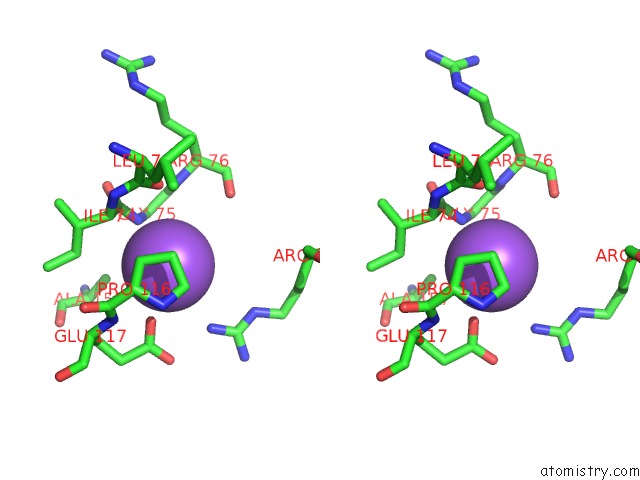
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Chaperone Complex within 5.0Å range:
|
Sodium binding site 2 out of 4 in 3cqx
Go back to
Sodium binding site 2 out
of 4 in the Chaperone Complex
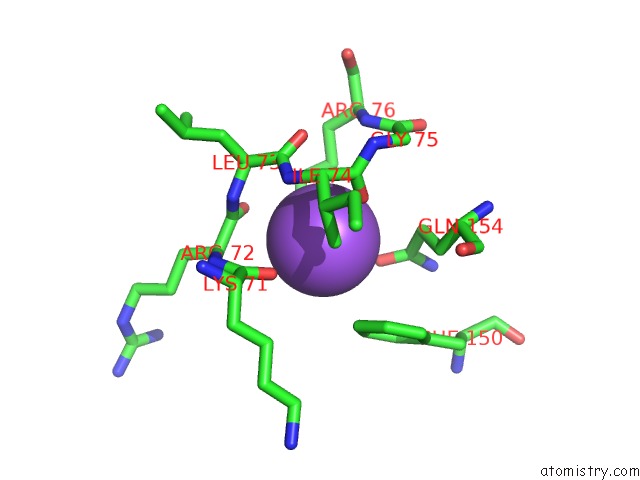
Mono view
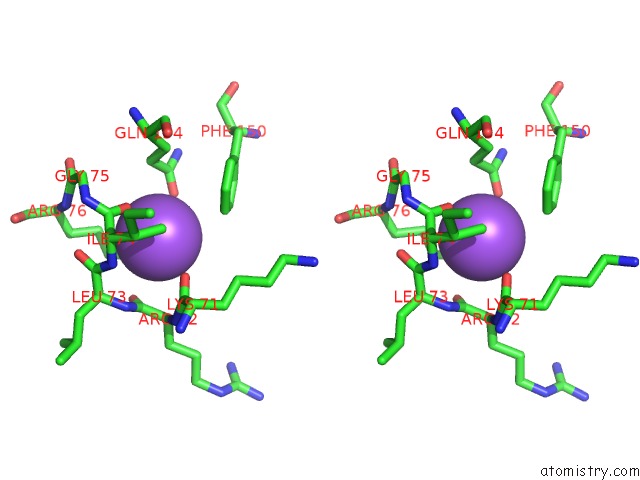
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Chaperone Complex within 5.0Å range:
|
Sodium binding site 3 out of 4 in 3cqx
Go back to
Sodium binding site 3 out
of 4 in the Chaperone Complex
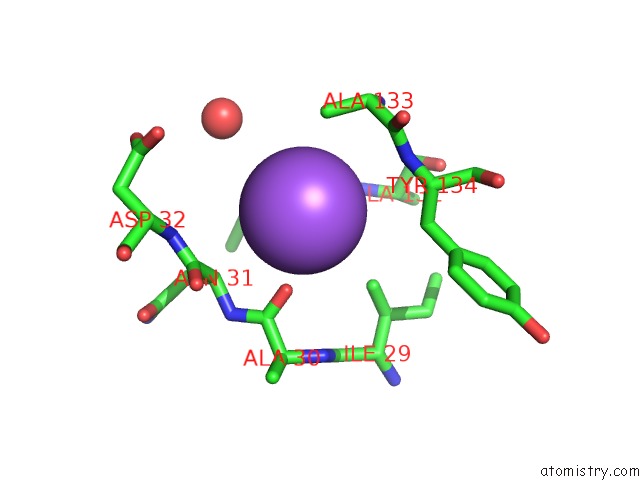
Mono view
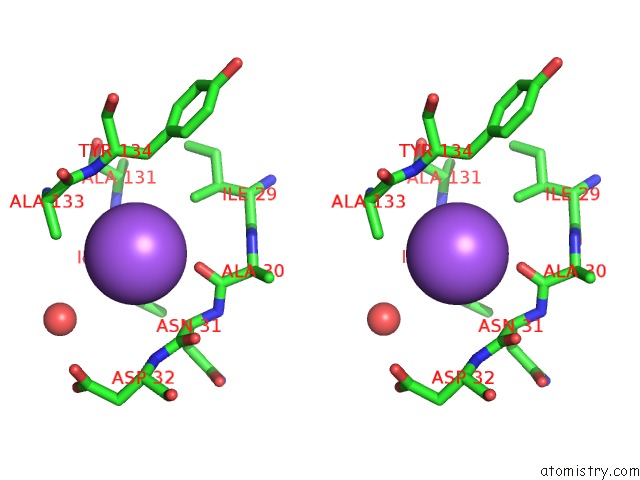
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 3 of Chaperone Complex within 5.0Å range:
|
Sodium binding site 4 out of 4 in 3cqx
Go back to
Sodium binding site 4 out
of 4 in the Chaperone Complex
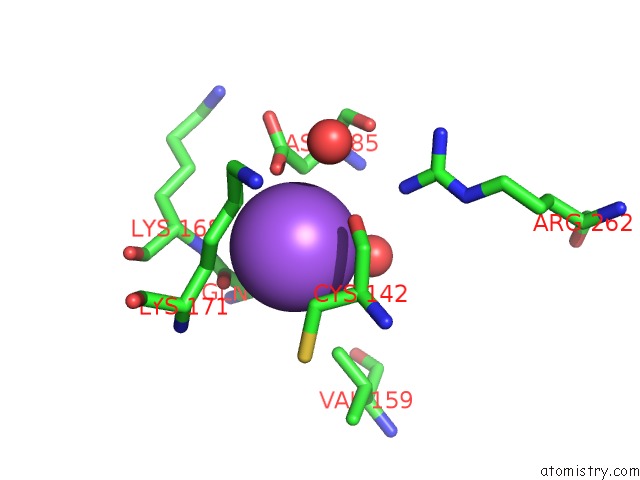
Mono view
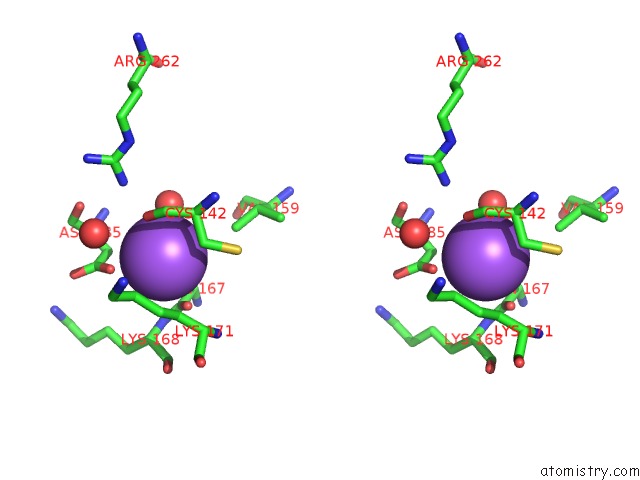
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 4 of Chaperone Complex within 5.0Å range:
|
Reference:
Z.Xu,
R.C.Page,
M.M.Gomes,
E.Kohli,
J.C.Nix,
A.B.Herr,
C.Patterson,
S.Misra.
Structural Basis of Nucleotide Exchange and Client Binding By the HSP70 Cochaperone BAG2 Nat.Struct.Mol.Biol. V. 15 1309 2008.
ISSN: ISSN 1545-9993
PubMed: 19029896
DOI: 10.1038/NSMB.1518
Page generated: Mon Oct 7 08:06:52 2024
ISSN: ISSN 1545-9993
PubMed: 19029896
DOI: 10.1038/NSMB.1518
Last articles
Ca in 2XHICa in 2XHJ
Ca in 2XHH
Ca in 2XF2
Ca in 2XFV
Ca in 2XFG
Ca in 2XCE
Ca in 2XFE
Ca in 2XFD
Ca in 2XEJ