Sodium »
PDB 2ok7-2ozt »
2ozt »
Sodium in PDB 2ozt: Crystal Structure of O-Succinylbenzoate Synthase From Thermosynechococcus Elongatus Bp-1
Protein crystallography data
The structure of Crystal Structure of O-Succinylbenzoate Synthase From Thermosynechococcus Elongatus Bp-1, PDB code: 2ozt
was solved by
V.N.Malashkevich,
J.Bonanno,
R.Toro,
J.M.Sauder,
K.D.Schwinn,
K.T.Bain,
J.M.Adams,
C.Reyes,
I.Rooney,
T.Gheyi,
S.R.Wasserman,
S.Emtage,
S.K.Burley,
S.C.Almo,
New York Sgx Research Center For Structuralgenomics (Nysgxrc),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.70 / 1.42 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 39.641, 113.498, 148.155, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.8 / 20.7 |
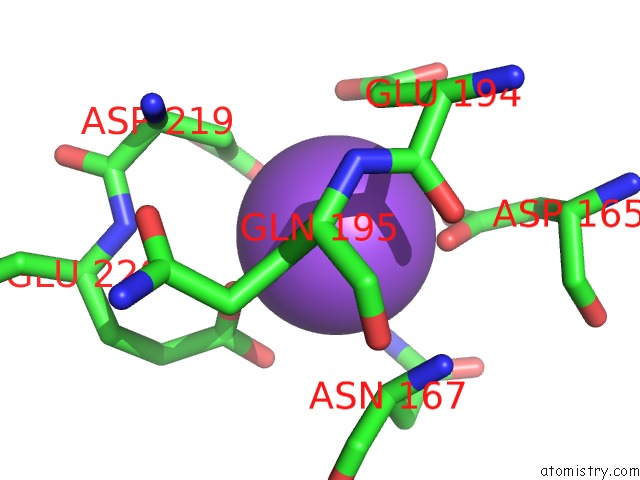
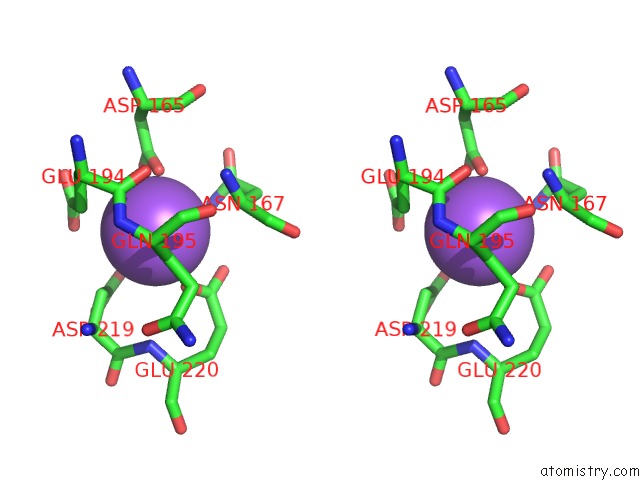
Sodium Binding Sites:
The binding sites of Sodium atom in the Crystal Structure of O-Succinylbenzoate Synthase From Thermosynechococcus Elongatus Bp-1
(pdb code 2ozt). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Crystal Structure of O-Succinylbenzoate Synthase From Thermosynechococcus Elongatus Bp-1, PDB code: 2ozt:
In total only one binding site of Sodium was determined in the Crystal Structure of O-Succinylbenzoate Synthase From Thermosynechococcus Elongatus Bp-1, PDB code: 2ozt:
Sodium binding site 1 out of 1 in 2ozt
Go back to
Sodium binding site 1 out
of 1 in the Crystal Structure of O-Succinylbenzoate Synthase From Thermosynechococcus Elongatus Bp-1
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Crystal Structure of O-Succinylbenzoate Synthase From Thermosynechococcus Elongatus Bp-1 within 5.0Å range:
|
Reference:
D.Odokonyero,
A.Sakai,
Y.Patskovsky,
V.N.Malashkevich,
A.A.Fedorov,
J.B.Bonanno,
E.V.Fedorov,
R.Toro,
R.Agarwal,
C.Wang,
N.D.Ozerova,
W.S.Yew,
J.M.Sauder,
S.Swaminathan,
S.K.Burley,
S.C.Almo,
M.E.Glasner.
Loss of Quaternary Structure Is Associated with Rapid Sequence Divergence in the Osbs Family. Proc.Natl.Acad.Sci.Usa V. 111 8535 2014.
ISSN: ISSN 0027-8424
PubMed: 24872444
DOI: 10.1073/PNAS.1318703111
Page generated: Mon Oct 7 03:21:18 2024
ISSN: ISSN 0027-8424
PubMed: 24872444
DOI: 10.1073/PNAS.1318703111
Last articles
Cl in 5FDJCl in 5FDL
Cl in 5FCZ
Cl in 5FDA
Cl in 5FDG
Cl in 5FDF
Cl in 5FDD
Cl in 5FCP
Cl in 5FCU
Cl in 5FCC