Sodium »
PDB 1v55-1w4n »
1w4n »
Sodium in PDB 1w4n: Agao Covalent Complex with Tranylcypromine
Enzymatic activity of Agao Covalent Complex with Tranylcypromine
All present enzymatic activity of Agao Covalent Complex with Tranylcypromine:
1.4.3.6;
1.4.3.6;
Protein crystallography data
The structure of Agao Covalent Complex with Tranylcypromine, PDB code: 1w4n
was solved by
A.P.Duff,
D.M.Trambaiolo,
D.B.Langley,
G.A.Juda,
E.M.Shepard,
D.M.Dooley,
H.C.Freeman,
J.M.Guss,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 28.28 / 1.65 |
| Space group | I 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 158.113, 63.001, 184.490, 90.00, 112.04, 90.00 |
| R / Rfree (%) | 17.1 / 19.9 |
Other elements in 1w4n:
The structure of Agao Covalent Complex with Tranylcypromine also contains other interesting chemical elements:
| Copper | (Cu) | 2 atoms |
Sodium Binding Sites:
The binding sites of Sodium atom in the Agao Covalent Complex with Tranylcypromine
(pdb code 1w4n). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 2 binding sites of Sodium where determined in the Agao Covalent Complex with Tranylcypromine, PDB code: 1w4n:
Jump to Sodium binding site number: 1; 2;
In total 2 binding sites of Sodium where determined in the Agao Covalent Complex with Tranylcypromine, PDB code: 1w4n:
Jump to Sodium binding site number: 1; 2;
Sodium binding site 1 out of 2 in 1w4n
Go back to
Sodium binding site 1 out
of 2 in the Agao Covalent Complex with Tranylcypromine
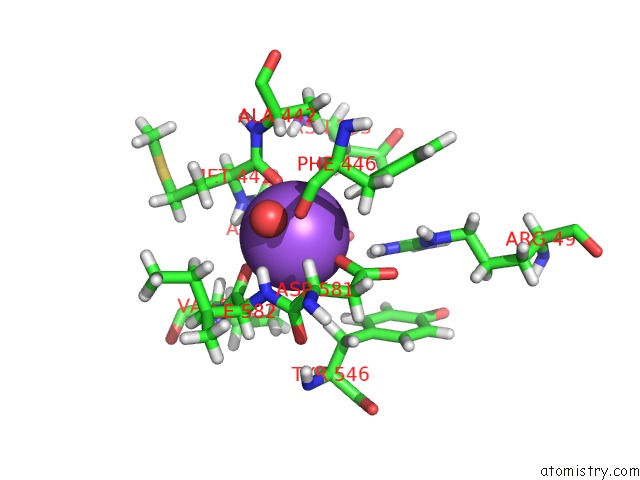
Mono view
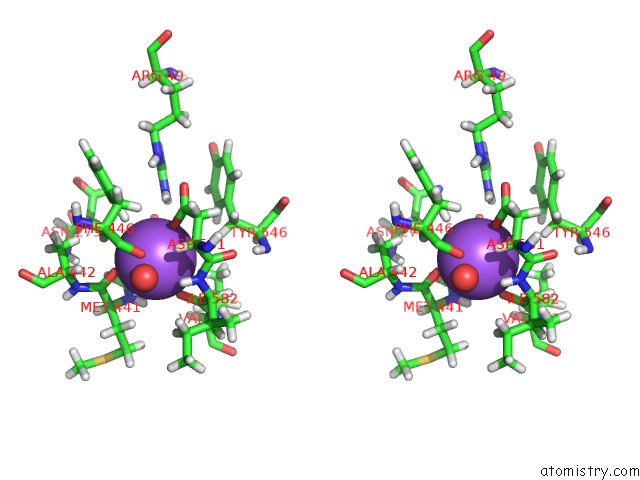
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Agao Covalent Complex with Tranylcypromine within 5.0Å range:
|
Sodium binding site 2 out of 2 in 1w4n
Go back to
Sodium binding site 2 out
of 2 in the Agao Covalent Complex with Tranylcypromine
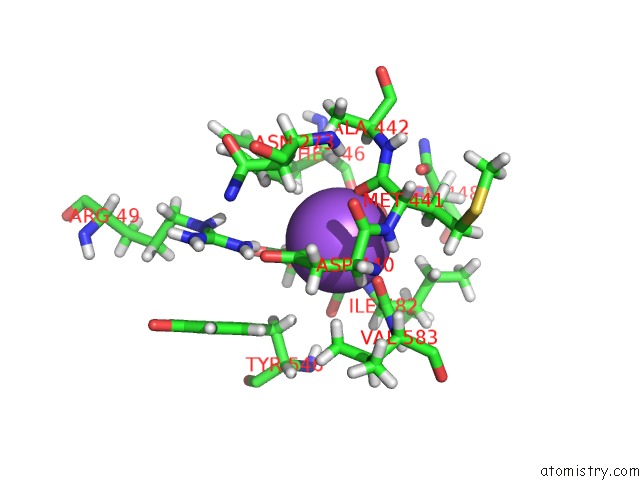
Mono view
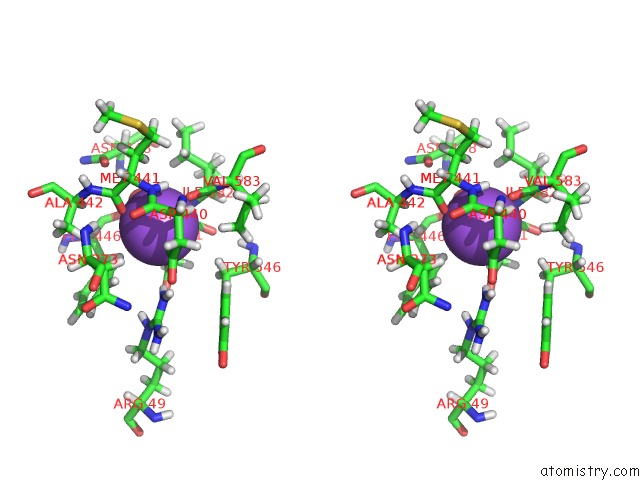
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Agao Covalent Complex with Tranylcypromine within 5.0Å range:
|
Reference:
D.B.Langley,
D.M.Trambaiolo,
A.P.Duff,
D.M.Dooley,
H.C.Freeman,
J.M.Guss.
Complexes of the Copper-Containing Amine Oxidase From Arthrobacter Globiformis with the Inhibitors Benzylhydrazine and Tranylcypromine. Acta Crystallogr.,Sect.F V. 64 577 2008.
ISSN: ESSN 1744-3091
PubMed: 18607080
DOI: 10.1107/S174430910801556X
Page generated: Sun Aug 17 08:59:04 2025
ISSN: ESSN 1744-3091
PubMed: 18607080
DOI: 10.1107/S174430910801556X
Last articles
Na in 8BGYNa in 8BGJ
Na in 8BDC
Na in 8BFD
Na in 8BB0
Na in 8BBS
Na in 8BBY
Na in 8BAZ
Na in 8BBU
Na in 8B8E