Sodium »
PDB 1ghy-1hn1 »
1h18 »
Sodium in PDB 1h18: Pyruvate Formate-Lyase (E.Coli) in Complex with Pyruvate
Enzymatic activity of Pyruvate Formate-Lyase (E.Coli) in Complex with Pyruvate
All present enzymatic activity of Pyruvate Formate-Lyase (E.Coli) in Complex with Pyruvate:
2.3.1.54;
2.3.1.54;
Protein crystallography data
The structure of Pyruvate Formate-Lyase (E.Coli) in Complex with Pyruvate, PDB code: 1h18
was solved by
A.Becker,
W.Kabsch,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 15 / 2.3 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 158.905, 158.905, 159.922, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 18.3 / 23.4 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Pyruvate Formate-Lyase (E.Coli) in Complex with Pyruvate
(pdb code 1h18). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 7 binding sites of Sodium where determined in the Pyruvate Formate-Lyase (E.Coli) in Complex with Pyruvate, PDB code: 1h18:
Jump to Sodium binding site number: 1; 2; 3; 4; 5; 6; 7;
In total 7 binding sites of Sodium where determined in the Pyruvate Formate-Lyase (E.Coli) in Complex with Pyruvate, PDB code: 1h18:
Jump to Sodium binding site number: 1; 2; 3; 4; 5; 6; 7;
Sodium binding site 1 out of 7 in 1h18
Go back to
Sodium binding site 1 out
of 7 in the Pyruvate Formate-Lyase (E.Coli) in Complex with Pyruvate
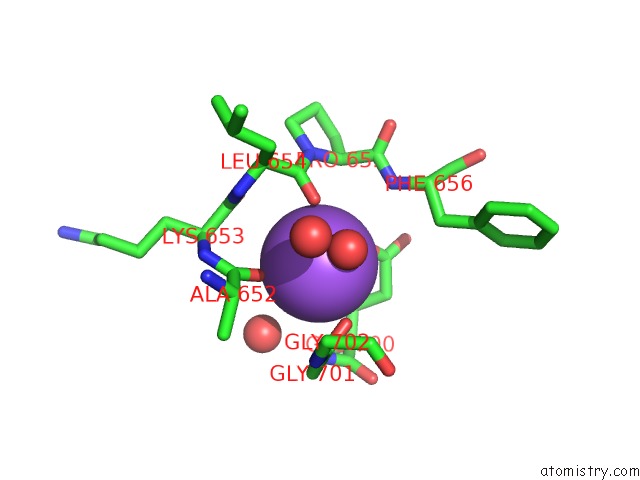
Mono view
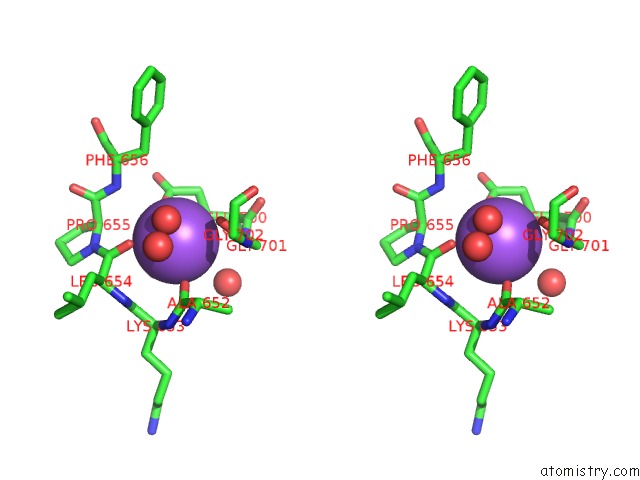
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Pyruvate Formate-Lyase (E.Coli) in Complex with Pyruvate within 5.0Å range:
|
Sodium binding site 2 out of 7 in 1h18
Go back to
Sodium binding site 2 out
of 7 in the Pyruvate Formate-Lyase (E.Coli) in Complex with Pyruvate
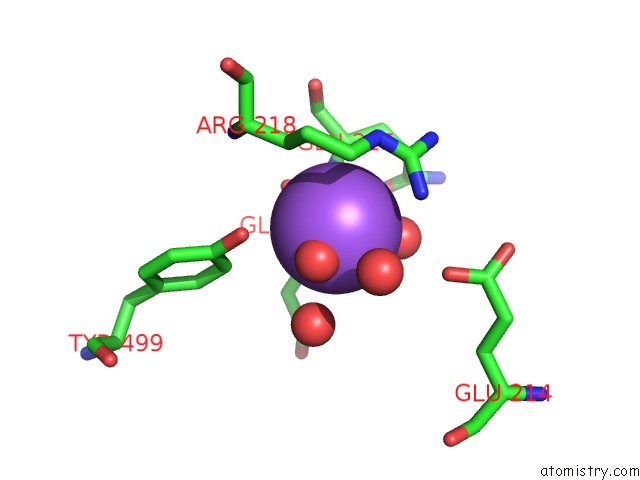
Mono view
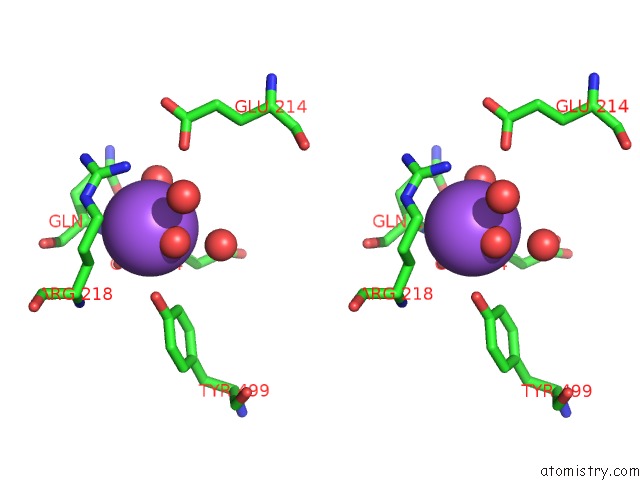
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Pyruvate Formate-Lyase (E.Coli) in Complex with Pyruvate within 5.0Å range:
|
Sodium binding site 3 out of 7 in 1h18
Go back to
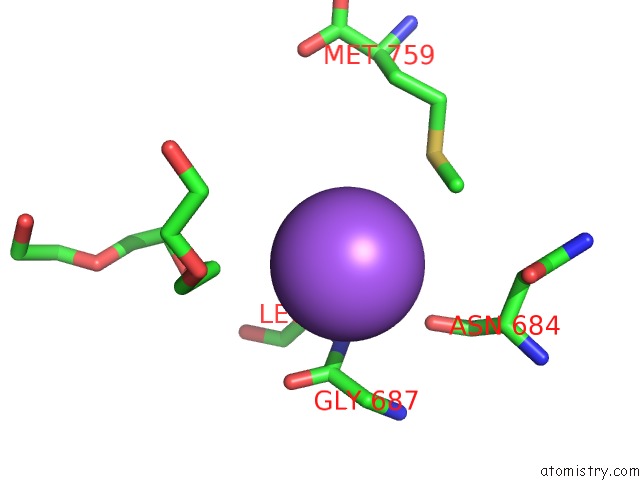
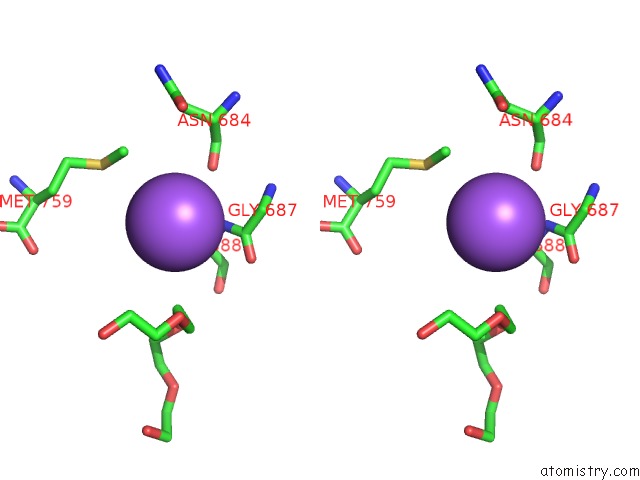
Sodium binding site 3 out
of 7 in the Pyruvate Formate-Lyase (E.Coli) in Complex with Pyruvate
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 3 of Pyruvate Formate-Lyase (E.Coli) in Complex with Pyruvate within 5.0Å range:
|
Sodium binding site 4 out of 7 in 1h18
Go back to
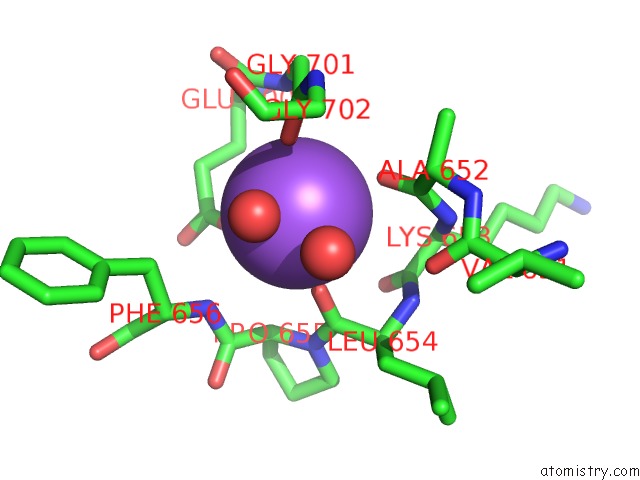
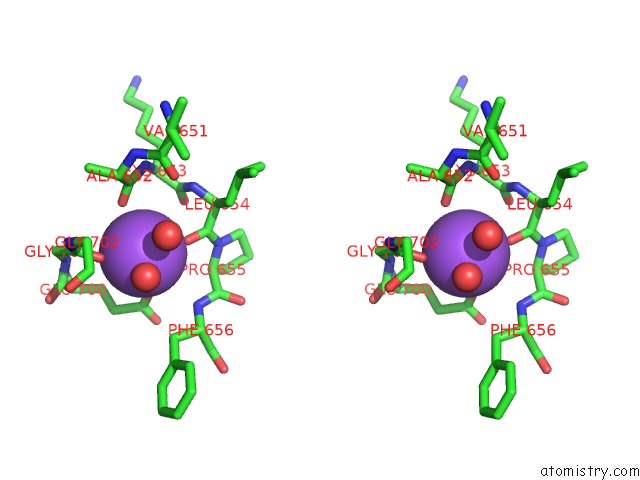
Sodium binding site 4 out
of 7 in the Pyruvate Formate-Lyase (E.Coli) in Complex with Pyruvate
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 4 of Pyruvate Formate-Lyase (E.Coli) in Complex with Pyruvate within 5.0Å range:
|
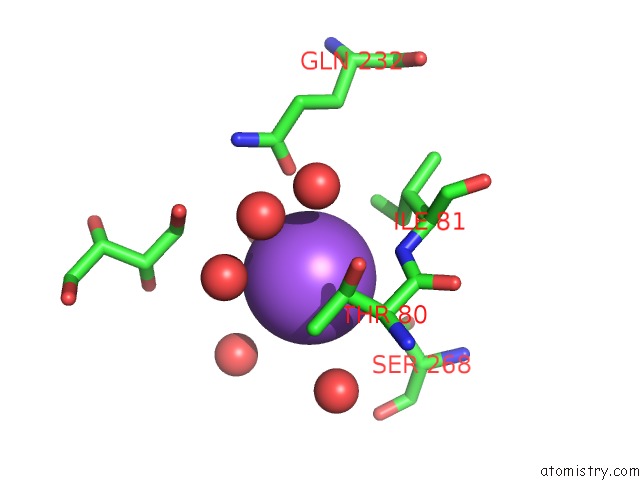
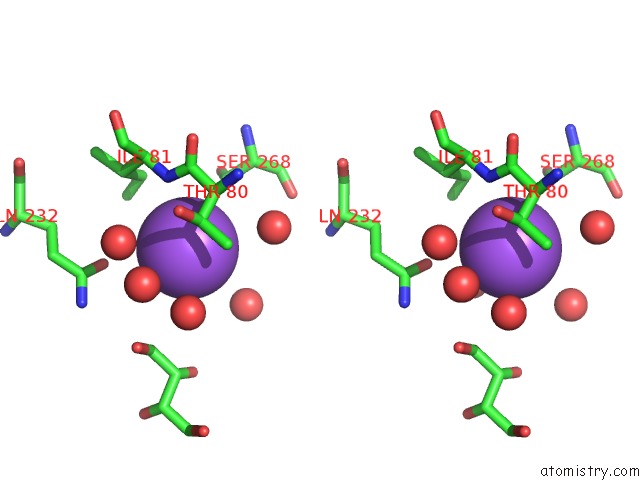
Sodium binding site 5 out of 7 in 1h18
Go back to
Sodium binding site 5 out
of 7 in the Pyruvate Formate-Lyase (E.Coli) in Complex with Pyruvate
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 5 of Pyruvate Formate-Lyase (E.Coli) in Complex with Pyruvate within 5.0Å range:
|
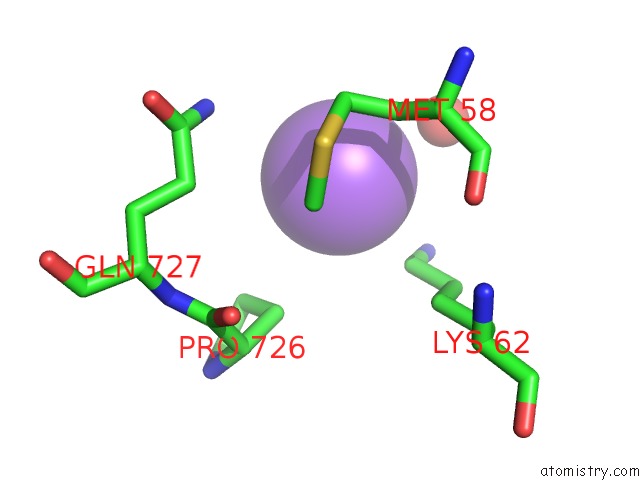
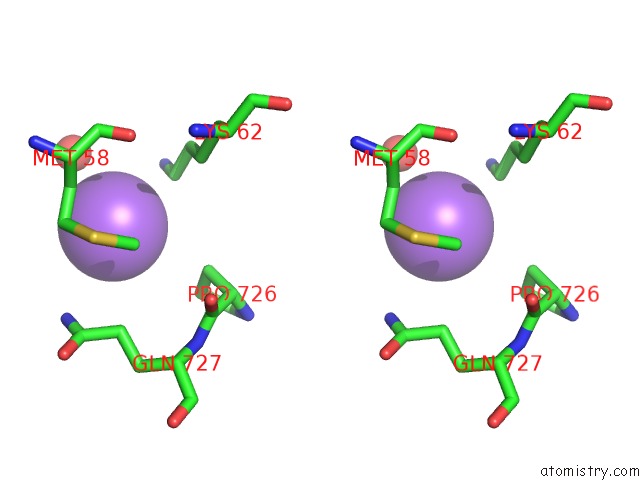
Sodium binding site 6 out of 7 in 1h18
Go back to
Sodium binding site 6 out
of 7 in the Pyruvate Formate-Lyase (E.Coli) in Complex with Pyruvate
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 6 of Pyruvate Formate-Lyase (E.Coli) in Complex with Pyruvate within 5.0Å range:
|
Sodium binding site 7 out of 7 in 1h18
Go back to
Sodium binding site 7 out
of 7 in the Pyruvate Formate-Lyase (E.Coli) in Complex with Pyruvate
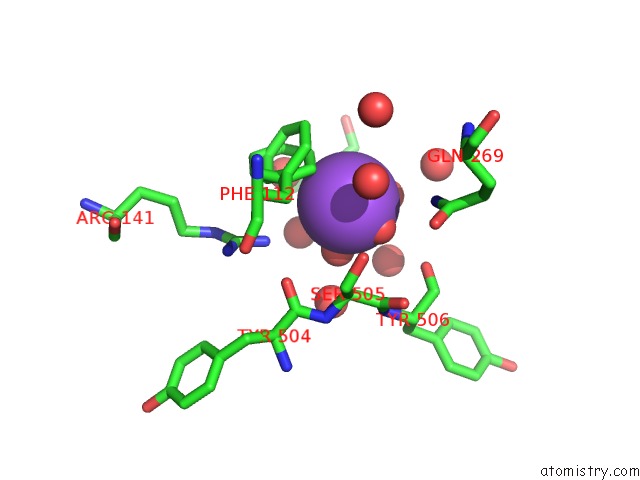
Mono view
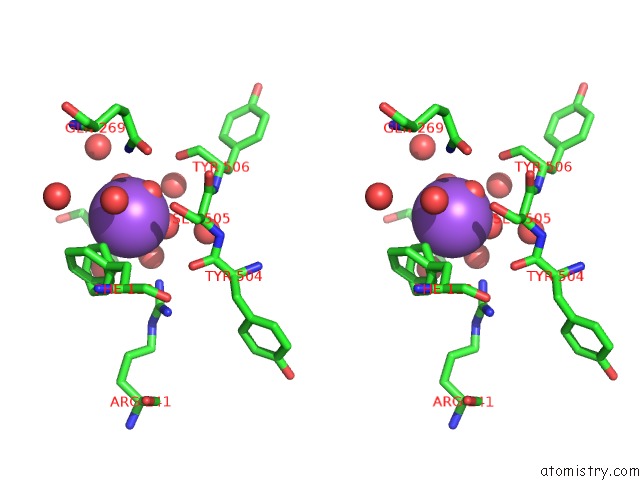
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 7 of Pyruvate Formate-Lyase (E.Coli) in Complex with Pyruvate within 5.0Å range:
|
Reference:
A.Becker,
W.Kabsch.
X-Ray Structure of Pyruvate Formate-Lyase in Complex with Pyruvate and Coa.How the Enzyme Uses the Cys-418 Thiyl Radical For Pyruvate Cleavage J.Biol.Chem. V. 277 40036 2002.
ISSN: ISSN 0021-9258
PubMed: 12163496
DOI: 10.1074/JBC.M205821200
Page generated: Sun Aug 17 05:08:50 2025
ISSN: ISSN 0021-9258
PubMed: 12163496
DOI: 10.1074/JBC.M205821200
Last articles
Na in 4OLFNa in 4OLN
Na in 4OHC
Na in 4OGL
Na in 4OJ1
Na in 4OFI
Na in 4OIC
Na in 4OF8
Na in 4OFX
Na in 4OE2