Sodium »
PDB 8c9g-8csf »
8can »
Sodium in PDB 8can: Cryo-Em Structure of the Cora Homohexamer From Galleria Mellonella Saliva
Other elements in 8can:
The structure of Cryo-Em Structure of the Cora Homohexamer From Galleria Mellonella Saliva also contains other interesting chemical elements:
| Copper | (Cu) | 24 atoms |
Sodium Binding Sites:
The binding sites of Sodium atom in the Cryo-Em Structure of the Cora Homohexamer From Galleria Mellonella Saliva
(pdb code 8can). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 3 binding sites of Sodium where determined in the Cryo-Em Structure of the Cora Homohexamer From Galleria Mellonella Saliva, PDB code: 8can:
Jump to Sodium binding site number: 1; 2; 3;
In total 3 binding sites of Sodium where determined in the Cryo-Em Structure of the Cora Homohexamer From Galleria Mellonella Saliva, PDB code: 8can:
Jump to Sodium binding site number: 1; 2; 3;
Sodium binding site 1 out of 3 in 8can
Go back to
Sodium binding site 1 out
of 3 in the Cryo-Em Structure of the Cora Homohexamer From Galleria Mellonella Saliva
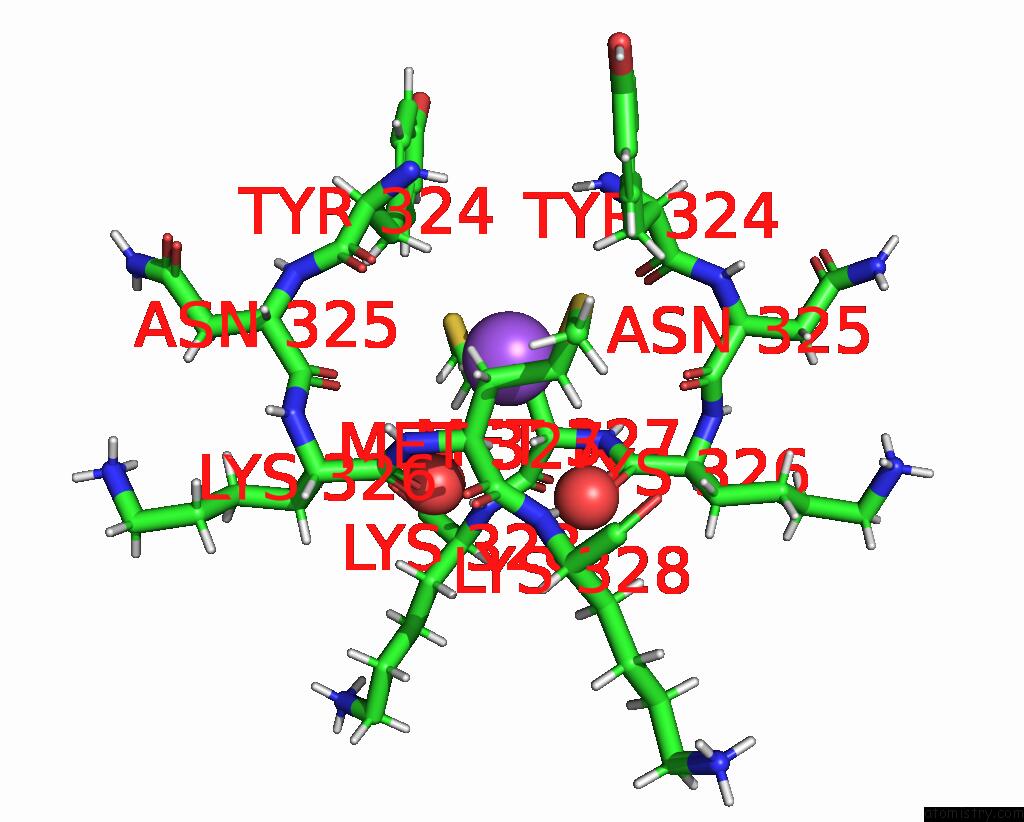
Mono view
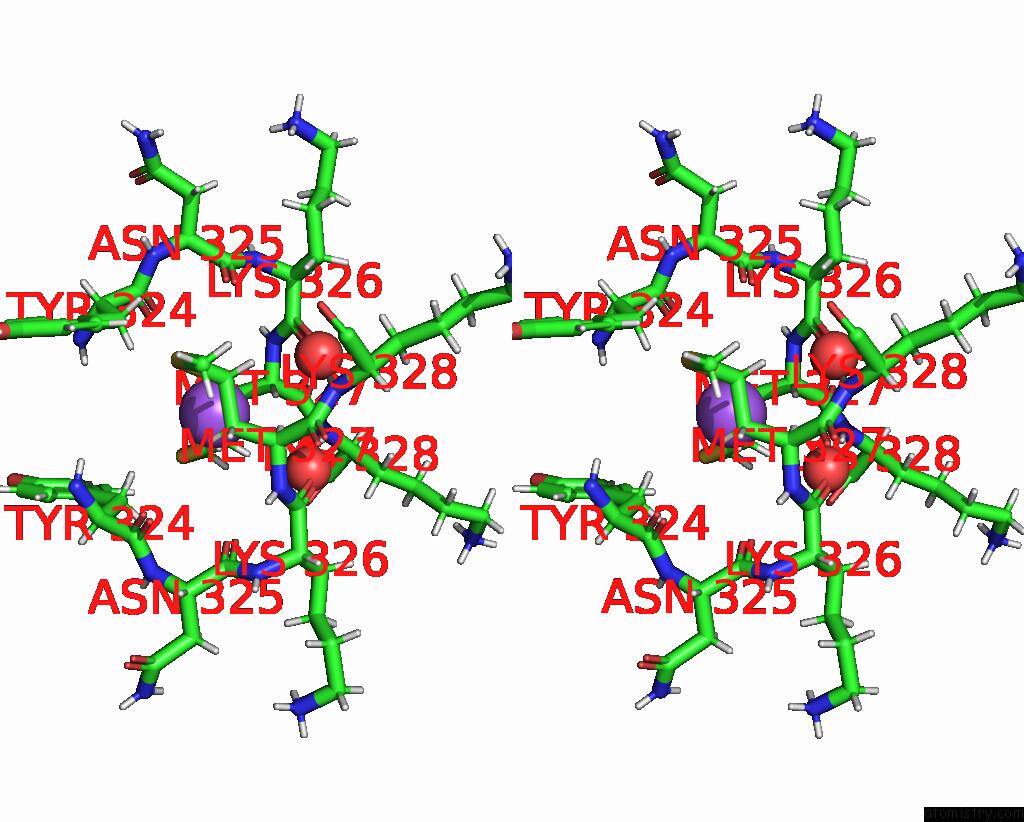
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Cryo-Em Structure of the Cora Homohexamer From Galleria Mellonella Saliva within 5.0Å range:
|
Sodium binding site 2 out of 3 in 8can
Go back to
Sodium binding site 2 out
of 3 in the Cryo-Em Structure of the Cora Homohexamer From Galleria Mellonella Saliva
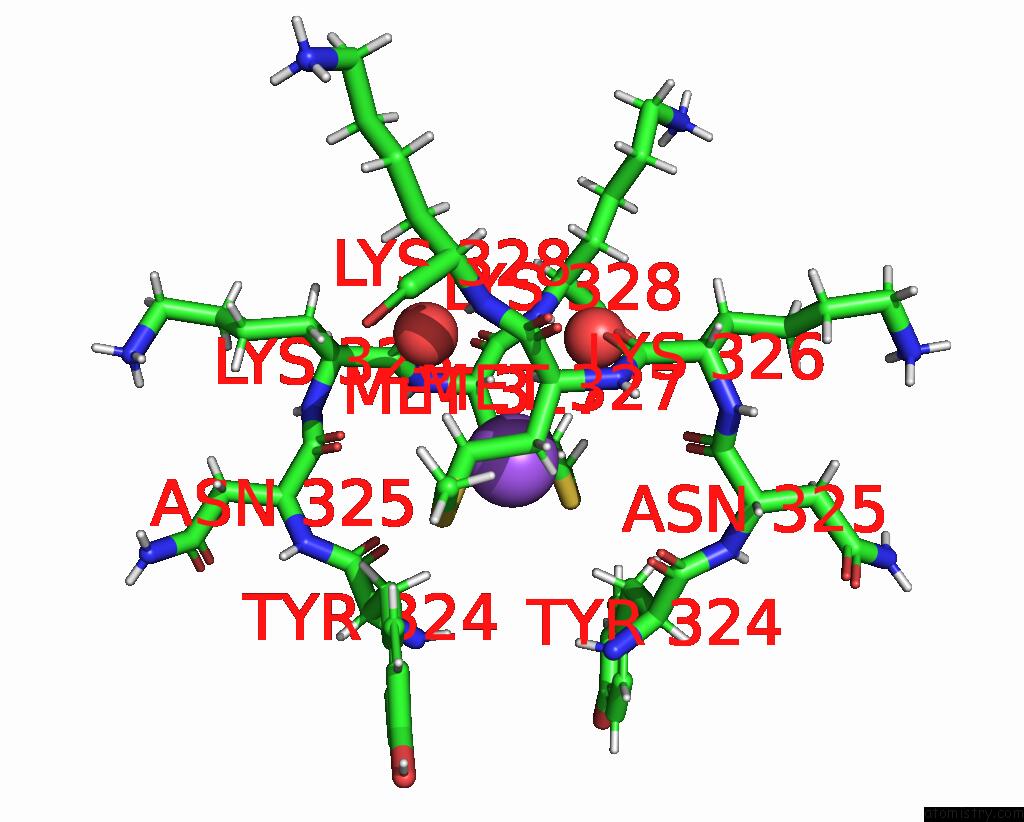
Mono view
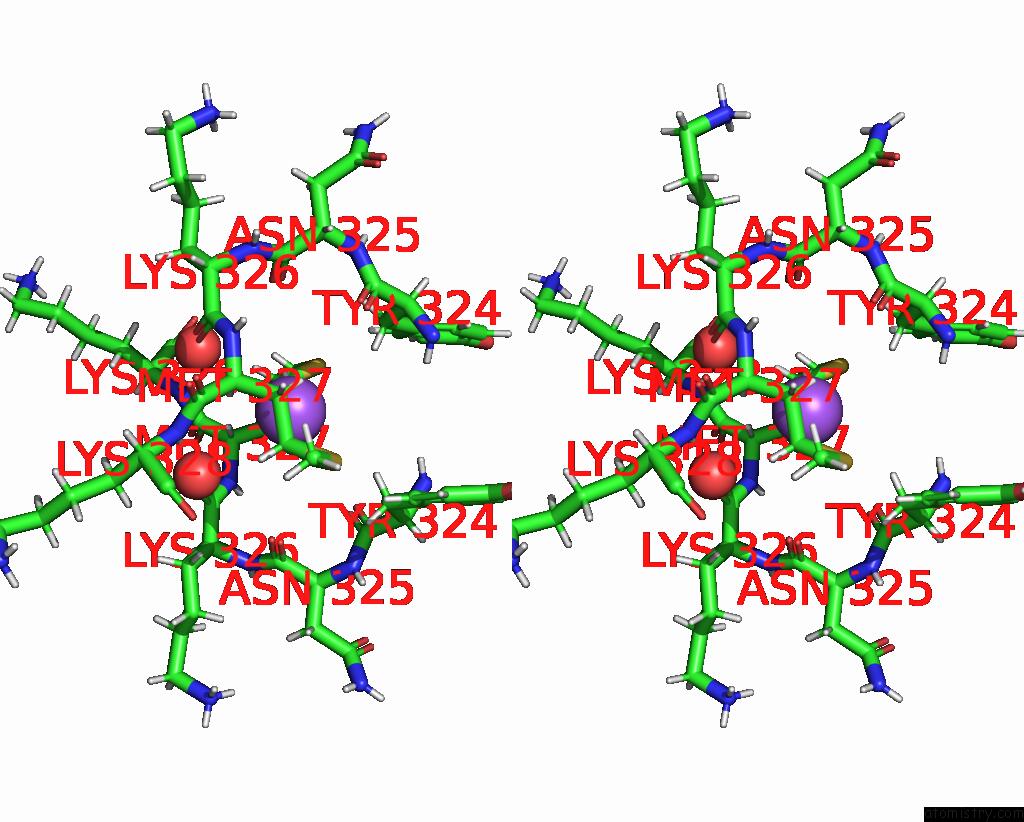
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Cryo-Em Structure of the Cora Homohexamer From Galleria Mellonella Saliva within 5.0Å range:
|
Sodium binding site 3 out of 3 in 8can
Go back to
Sodium binding site 3 out
of 3 in the Cryo-Em Structure of the Cora Homohexamer From Galleria Mellonella Saliva
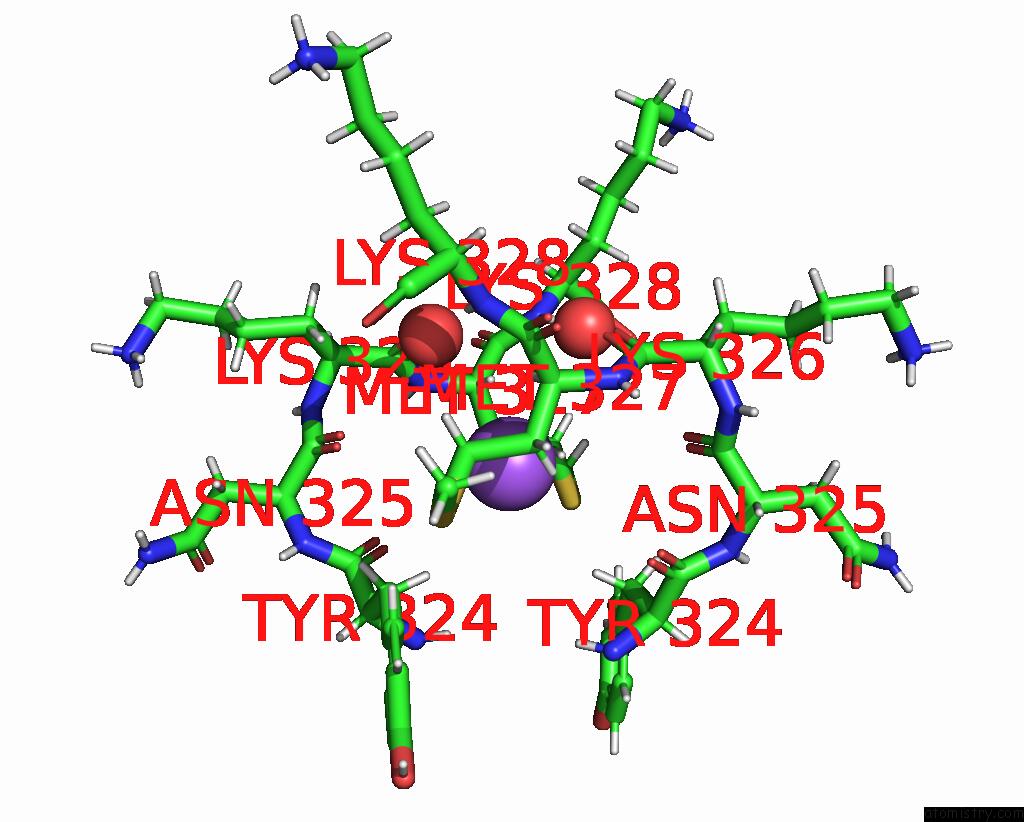
Mono view
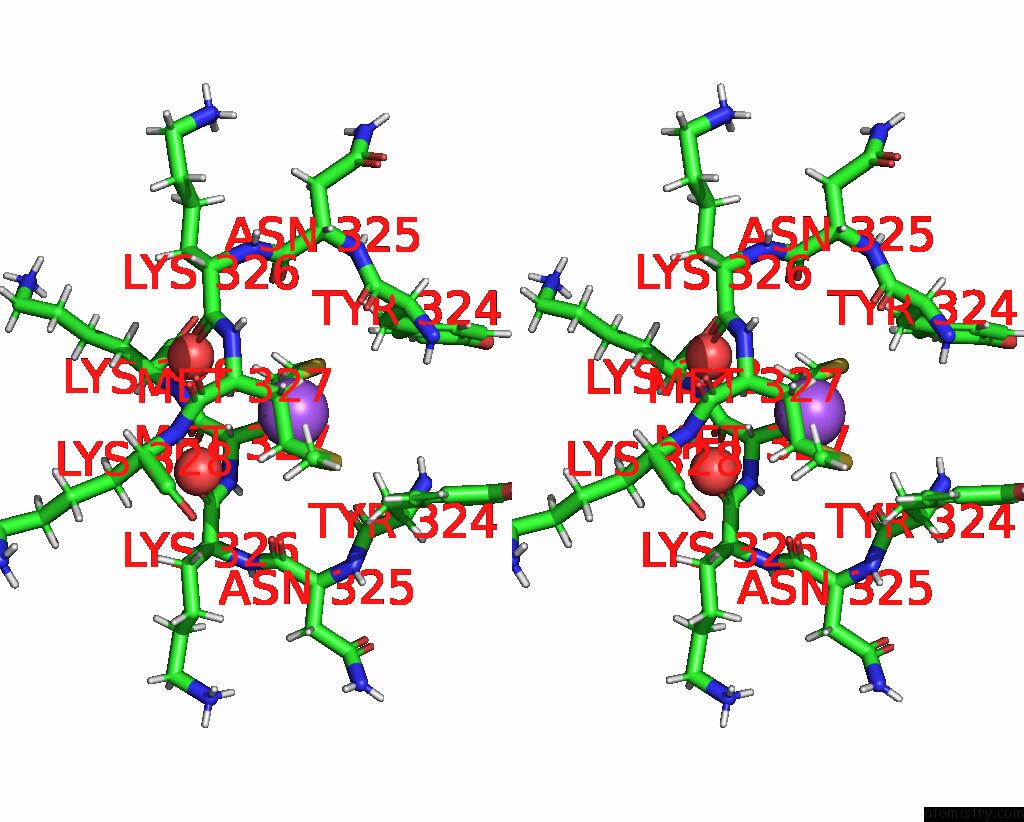
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 3 of Cryo-Em Structure of the Cora Homohexamer From Galleria Mellonella Saliva within 5.0Å range:
|
Reference:
M.Spinola-Amilibia,
R.Illanes-Vicioso,
E.Ruiz-L Formula See Text Pez,
P.Colomer-Vidal,
F.V.Rodriguez,
R.Peces Perez,
C.F.Arias,
T.Torroba,
M.Sol Formula See Text,
E.Arias-Palomo,
F.Bertocchini.
Plastic Degradation By Insect Hexamerins: Near-Atomic Resolution Structures of the Polyethylene-Degrading Proteins From the Wax Worm Saliva. Sci Adv V. 9 I6813 2023.
ISSN: ESSN 2375-2548
PubMed: 37729416
DOI: 10.1126/SCIADV.ADI6813
Page generated: Mon Aug 18 13:25:35 2025
ISSN: ESSN 2375-2548
PubMed: 37729416
DOI: 10.1126/SCIADV.ADI6813
Last articles
Mn in 9LJUMn in 9LJW
Mn in 9LJS
Mn in 9LJR
Mn in 9LJT
Mn in 9LJV
Mg in 9UA2
Mg in 9R96
Mg in 9VM1
Mg in 9P01