Sodium »
PDB 6qu9-6r7r »
6r6u »
Sodium in PDB 6r6u: Crystal Structure of Human Cis-Aconitate Decarboxylase
Enzymatic activity of Crystal Structure of Human Cis-Aconitate Decarboxylase
All present enzymatic activity of Crystal Structure of Human Cis-Aconitate Decarboxylase:
4.1.1.6;
4.1.1.6;
Protein crystallography data
The structure of Crystal Structure of Human Cis-Aconitate Decarboxylase, PDB code: 6r6u
was solved by
P.Lukat,
F.Chen,
K.Saile,
K.Buessow,
F.Pessler,
W.Blankenfeldt,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 60.41 / 1.71 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 102.010, 109.980, 74.974, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 14.5 / 16.2 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Crystal Structure of Human Cis-Aconitate Decarboxylase
(pdb code 6r6u). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 6 binding sites of Sodium where determined in the Crystal Structure of Human Cis-Aconitate Decarboxylase, PDB code: 6r6u:
Jump to Sodium binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Sodium where determined in the Crystal Structure of Human Cis-Aconitate Decarboxylase, PDB code: 6r6u:
Jump to Sodium binding site number: 1; 2; 3; 4; 5; 6;
Sodium binding site 1 out of 6 in 6r6u
Go back to
Sodium binding site 1 out
of 6 in the Crystal Structure of Human Cis-Aconitate Decarboxylase
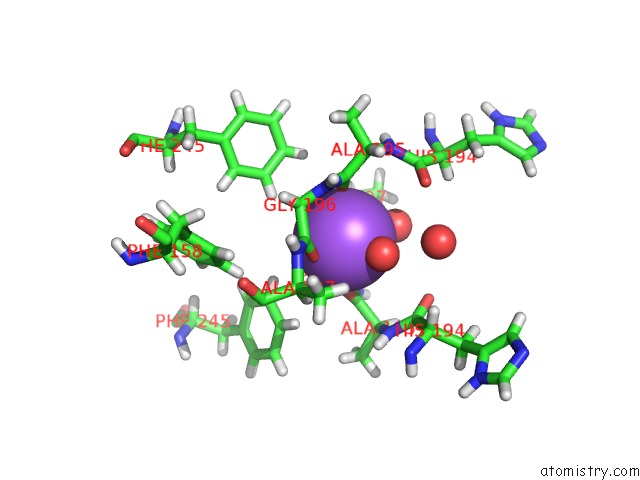
Mono view
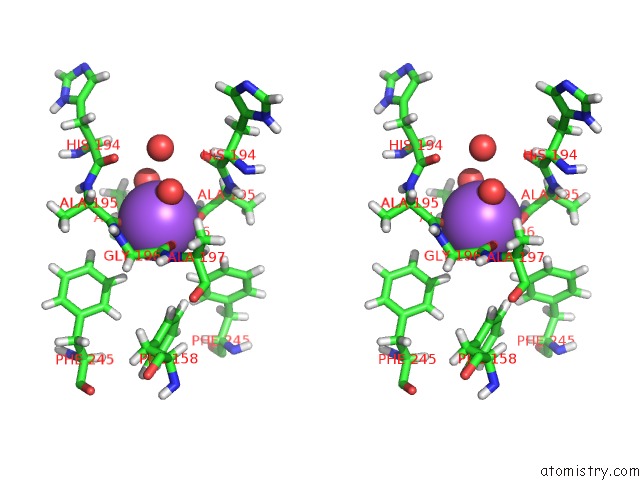
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Crystal Structure of Human Cis-Aconitate Decarboxylase within 5.0Å range:
|
Sodium binding site 2 out of 6 in 6r6u
Go back to
Sodium binding site 2 out
of 6 in the Crystal Structure of Human Cis-Aconitate Decarboxylase
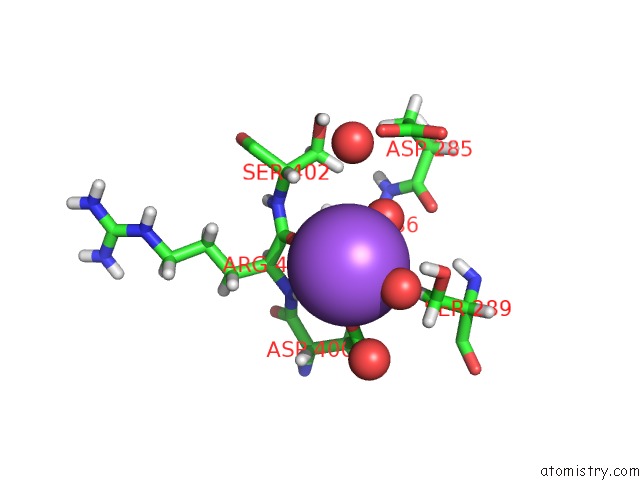
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Crystal Structure of Human Cis-Aconitate Decarboxylase within 5.0Å range:
|
Sodium binding site 3 out of 6 in 6r6u
Go back to
Sodium binding site 3 out
of 6 in the Crystal Structure of Human Cis-Aconitate Decarboxylase
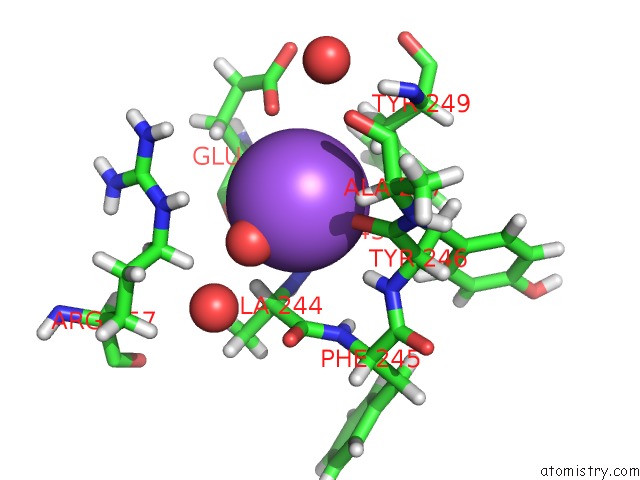
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 3 of Crystal Structure of Human Cis-Aconitate Decarboxylase within 5.0Å range:
|
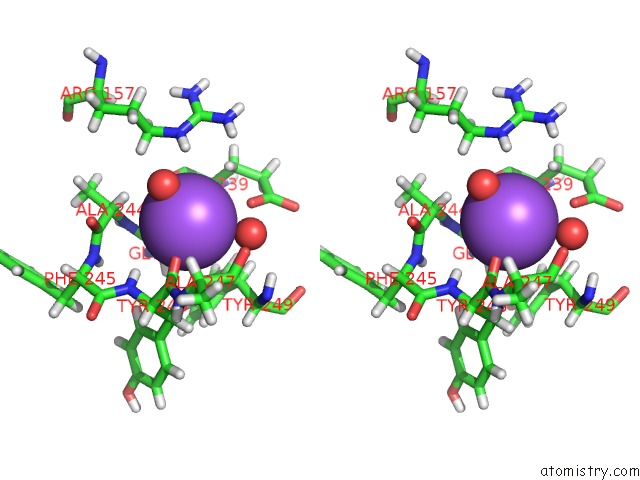
Sodium binding site 4 out of 6 in 6r6u
Go back to
Sodium binding site 4 out
of 6 in the Crystal Structure of Human Cis-Aconitate Decarboxylase
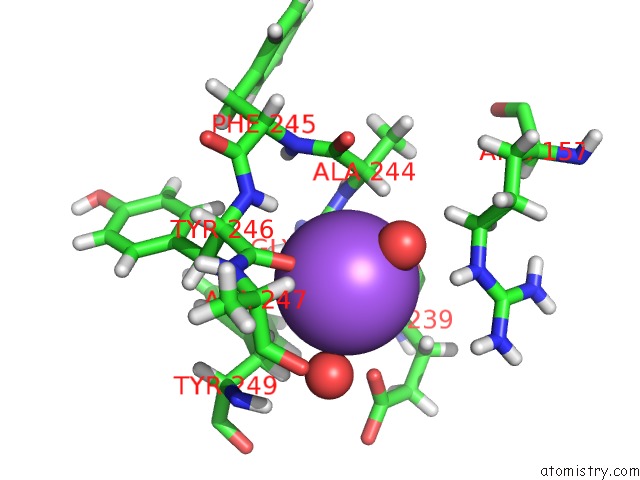
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 4 of Crystal Structure of Human Cis-Aconitate Decarboxylase within 5.0Å range:
|
Sodium binding site 5 out of 6 in 6r6u
Go back to
Sodium binding site 5 out
of 6 in the Crystal Structure of Human Cis-Aconitate Decarboxylase

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 5 of Crystal Structure of Human Cis-Aconitate Decarboxylase within 5.0Å range:
|
Sodium binding site 6 out of 6 in 6r6u
Go back to
Sodium binding site 6 out
of 6 in the Crystal Structure of Human Cis-Aconitate Decarboxylase
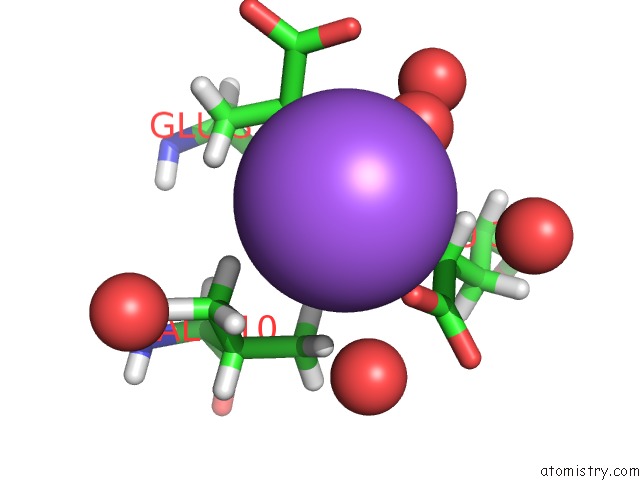
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 6 of Crystal Structure of Human Cis-Aconitate Decarboxylase within 5.0Å range:
|
Reference:
F.Chen,
P.Lukat,
A.A.Iqbal,
K.Saile,
V.Kaever,
J.Van Den Heuvel,
W.Blankenfeldt,
K.Bussow,
F.Pessler.
Crystal Structure Ofcis-Aconitate Decarboxylase Reveals the Impact of Naturally Occurring Human Mutations on Itaconate Synthesis. Proc.Natl.Acad.Sci.Usa V. 116 20644 2019.
ISSN: ESSN 1091-6490
PubMed: 31548418
DOI: 10.1073/PNAS.1908770116
Page generated: Mon Aug 18 06:52:40 2025
ISSN: ESSN 1091-6490
PubMed: 31548418
DOI: 10.1073/PNAS.1908770116
Last articles
Zn in 3HPAZn in 3HOV
Zn in 3HOX
Zn in 3HOW
Zn in 3HNA
Zn in 3HNJ
Zn in 3HNI
Zn in 3HO5
Zn in 3HN8
Zn in 3HM3