Sodium »
PDB 5mft-5mse »
5mft »
Sodium in PDB 5mft: The Crystal Structure of E. Coli Aminopeptidase N in Complex with 7- Amino-1-Bromo-4-Phenyl-5,7,8,9-Tetrahydrobenzocyclohepten-6-One
Enzymatic activity of The Crystal Structure of E. Coli Aminopeptidase N in Complex with 7- Amino-1-Bromo-4-Phenyl-5,7,8,9-Tetrahydrobenzocyclohepten-6-One
All present enzymatic activity of The Crystal Structure of E. Coli Aminopeptidase N in Complex with 7- Amino-1-Bromo-4-Phenyl-5,7,8,9-Tetrahydrobenzocyclohepten-6-One:
3.4.11.2;
3.4.11.2;
Protein crystallography data
The structure of The Crystal Structure of E. Coli Aminopeptidase N in Complex with 7- Amino-1-Bromo-4-Phenyl-5,7,8,9-Tetrahydrobenzocyclohepten-6-One, PDB code: 5mft
was solved by
G.Peng,
V.Olieric,
A.G.Mcewen,
C.Schmitt,
S.Albrecht,
J.Cavarelli,
C.Tarnus,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 60.33 / 1.59 |
| Space group | P 31 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 120.651, 120.651, 170.676, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 11.1 / 13.9 |
Other elements in 5mft:
The structure of The Crystal Structure of E. Coli Aminopeptidase N in Complex with 7- Amino-1-Bromo-4-Phenyl-5,7,8,9-Tetrahydrobenzocyclohepten-6-One also contains other interesting chemical elements:
| Zinc | (Zn) | 1 atom |
| Bromine | (Br) | 1 atom |
| Chlorine | (Cl) | 2 atoms |
Sodium Binding Sites:
The binding sites of Sodium atom in the The Crystal Structure of E. Coli Aminopeptidase N in Complex with 7- Amino-1-Bromo-4-Phenyl-5,7,8,9-Tetrahydrobenzocyclohepten-6-One
(pdb code 5mft). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 6 binding sites of Sodium where determined in the The Crystal Structure of E. Coli Aminopeptidase N in Complex with 7- Amino-1-Bromo-4-Phenyl-5,7,8,9-Tetrahydrobenzocyclohepten-6-One, PDB code: 5mft:
Jump to Sodium binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Sodium where determined in the The Crystal Structure of E. Coli Aminopeptidase N in Complex with 7- Amino-1-Bromo-4-Phenyl-5,7,8,9-Tetrahydrobenzocyclohepten-6-One, PDB code: 5mft:
Jump to Sodium binding site number: 1; 2; 3; 4; 5; 6;
Sodium binding site 1 out of 6 in 5mft
Go back to
Sodium binding site 1 out
of 6 in the The Crystal Structure of E. Coli Aminopeptidase N in Complex with 7- Amino-1-Bromo-4-Phenyl-5,7,8,9-Tetrahydrobenzocyclohepten-6-One
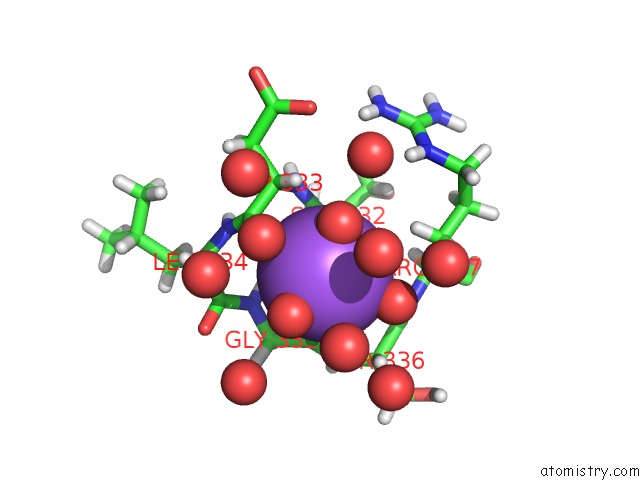
Mono view
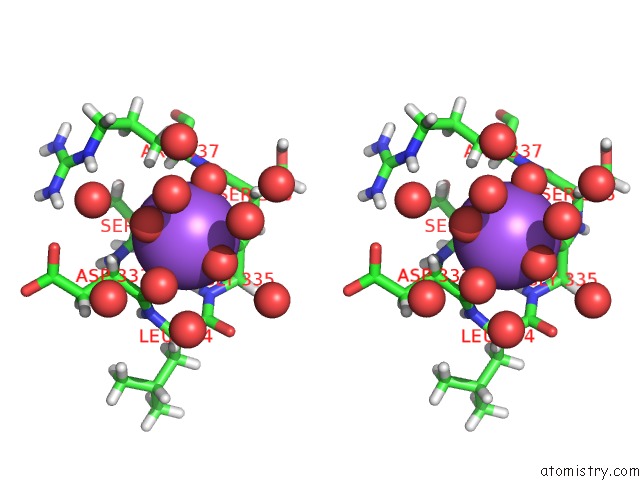
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of The Crystal Structure of E. Coli Aminopeptidase N in Complex with 7- Amino-1-Bromo-4-Phenyl-5,7,8,9-Tetrahydrobenzocyclohepten-6-One within 5.0Å range:
|
Sodium binding site 2 out of 6 in 5mft
Go back to
Sodium binding site 2 out
of 6 in the The Crystal Structure of E. Coli Aminopeptidase N in Complex with 7- Amino-1-Bromo-4-Phenyl-5,7,8,9-Tetrahydrobenzocyclohepten-6-One
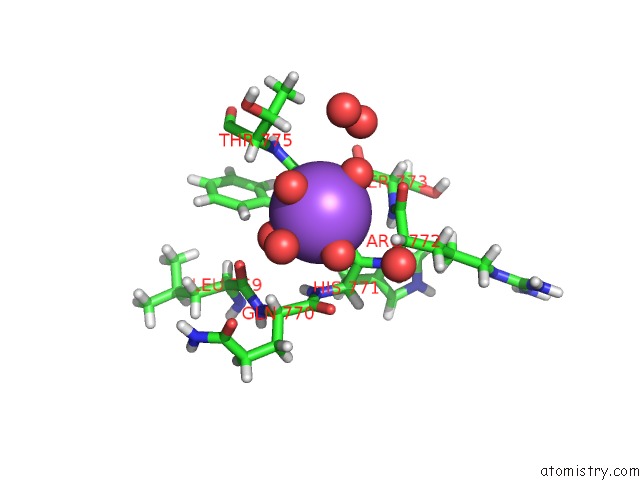
Mono view
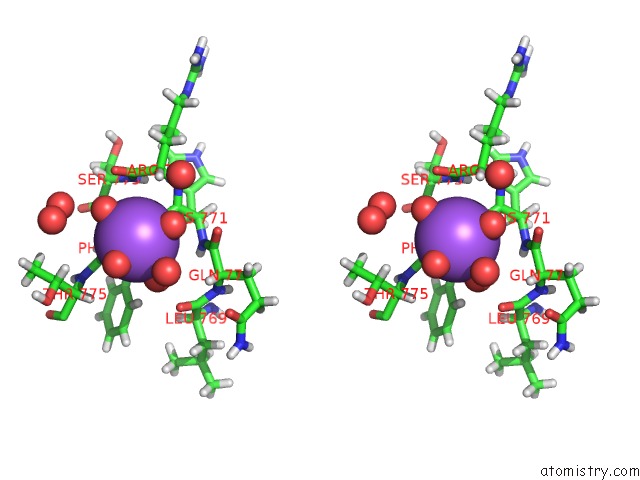
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of The Crystal Structure of E. Coli Aminopeptidase N in Complex with 7- Amino-1-Bromo-4-Phenyl-5,7,8,9-Tetrahydrobenzocyclohepten-6-One within 5.0Å range:
|
Sodium binding site 3 out of 6 in 5mft
Go back to
Sodium binding site 3 out
of 6 in the The Crystal Structure of E. Coli Aminopeptidase N in Complex with 7- Amino-1-Bromo-4-Phenyl-5,7,8,9-Tetrahydrobenzocyclohepten-6-One
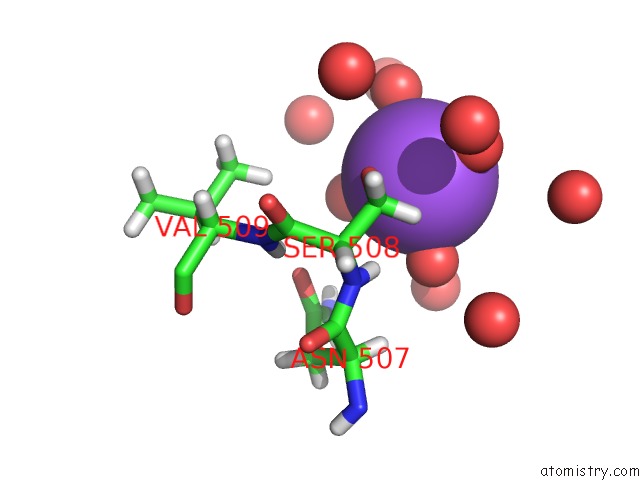
Mono view
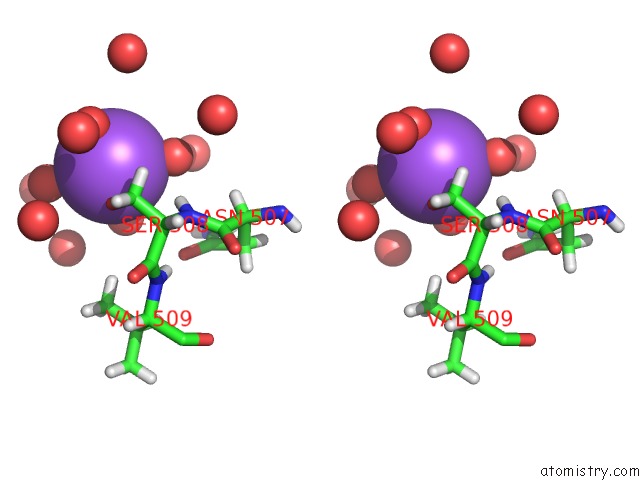
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 3 of The Crystal Structure of E. Coli Aminopeptidase N in Complex with 7- Amino-1-Bromo-4-Phenyl-5,7,8,9-Tetrahydrobenzocyclohepten-6-One within 5.0Å range:
|
Sodium binding site 4 out of 6 in 5mft
Go back to
Sodium binding site 4 out
of 6 in the The Crystal Structure of E. Coli Aminopeptidase N in Complex with 7- Amino-1-Bromo-4-Phenyl-5,7,8,9-Tetrahydrobenzocyclohepten-6-One
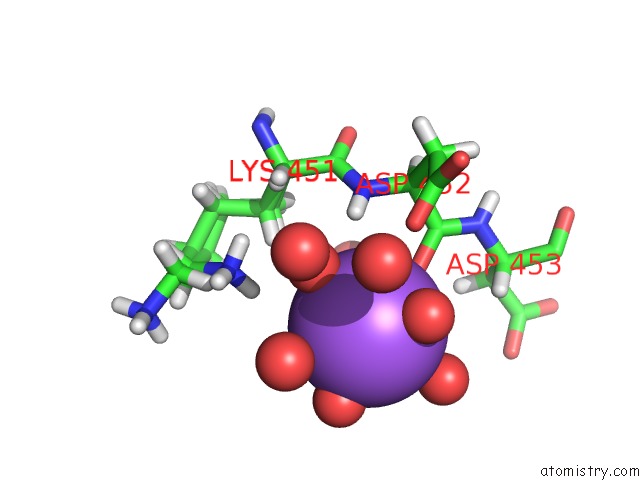
Mono view
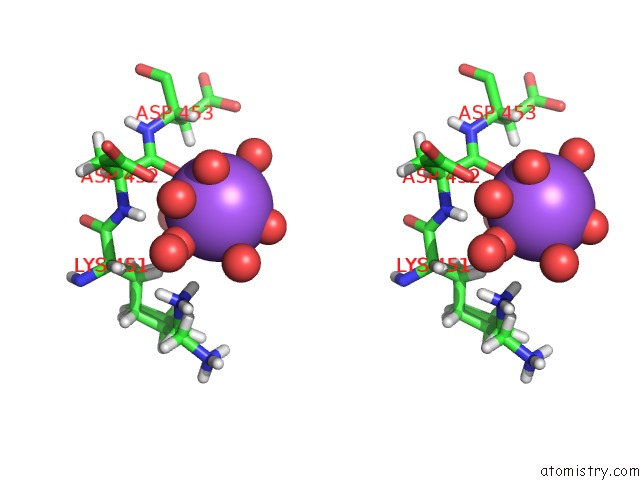
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 4 of The Crystal Structure of E. Coli Aminopeptidase N in Complex with 7- Amino-1-Bromo-4-Phenyl-5,7,8,9-Tetrahydrobenzocyclohepten-6-One within 5.0Å range:
|
Sodium binding site 5 out of 6 in 5mft
Go back to
Sodium binding site 5 out
of 6 in the The Crystal Structure of E. Coli Aminopeptidase N in Complex with 7- Amino-1-Bromo-4-Phenyl-5,7,8,9-Tetrahydrobenzocyclohepten-6-One
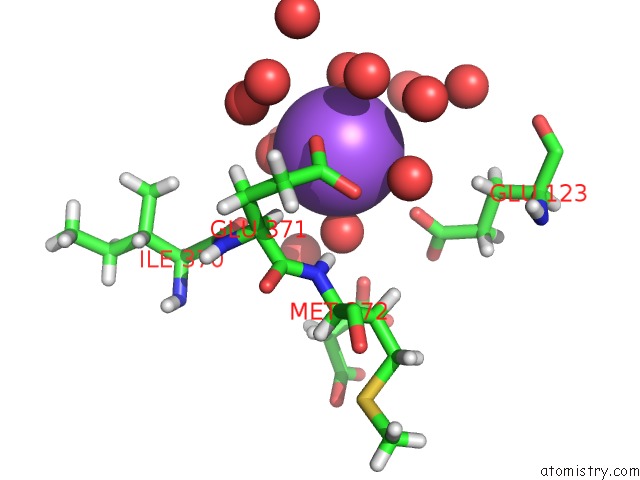
Mono view
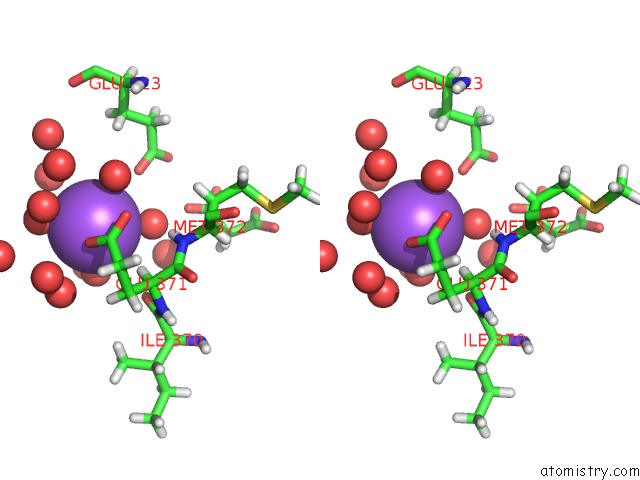
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 5 of The Crystal Structure of E. Coli Aminopeptidase N in Complex with 7- Amino-1-Bromo-4-Phenyl-5,7,8,9-Tetrahydrobenzocyclohepten-6-One within 5.0Å range:
|
Sodium binding site 6 out of 6 in 5mft
Go back to
Sodium binding site 6 out
of 6 in the The Crystal Structure of E. Coli Aminopeptidase N in Complex with 7- Amino-1-Bromo-4-Phenyl-5,7,8,9-Tetrahydrobenzocyclohepten-6-One
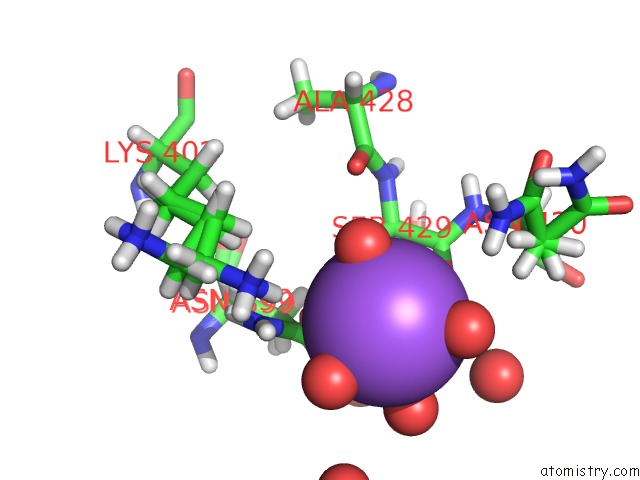
Mono view
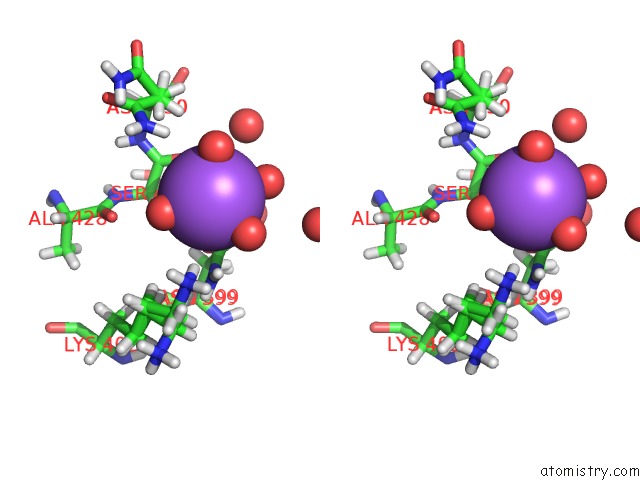
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 6 of The Crystal Structure of E. Coli Aminopeptidase N in Complex with 7- Amino-1-Bromo-4-Phenyl-5,7,8,9-Tetrahydrobenzocyclohepten-6-One within 5.0Å range:
|
Reference:
G.Peng,
A.G.Mcewen,
V.Olieric,
C.Schmitt,
S.Albrecht,
J.Cavarelli,
C.Tarnus.
Insight Into the Remarkable Affinity and Selectivity of the Aminobenzosuberone Scaffold For the M1 Aminopeptidases Family Based on Structure Analysis. Proteins V. 85 1413 2017.
ISSN: ESSN 1097-0134
PubMed: 28383176
DOI: 10.1002/PROT.25301
Page generated: Mon Oct 7 22:39:53 2024
ISSN: ESSN 1097-0134
PubMed: 28383176
DOI: 10.1002/PROT.25301
Last articles
K in 7NPXK in 7NWD
K in 7NXF
K in 7NF4
K in 7NHT
K in 7NKG
K in 7NF2
K in 7N9L
K in 7NFZ
K in 7NF3