Sodium »
PDB 5f01-5fjc »
5f7k »
Sodium in PDB 5f7k: Blood Group Antigen Binding Adhesin Baba of Helicobacter Pylori Strain 17875 in Complex with Nanobody Nb-ER19
Protein crystallography data
The structure of Blood Group Antigen Binding Adhesin Baba of Helicobacter Pylori Strain 17875 in Complex with Nanobody Nb-ER19, PDB code: 5f7k
was solved by
K.Moonens,
P.Gideonsson,
S.Subedi,
E.Romao,
S.Oscarson,
S.Muyldermans,
T.Boren,
H.Remaut,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 58.04 / 2.17 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 50.990, 131.660, 123.460, 90.00, 94.77, 90.00 |
| R / Rfree (%) | 17.1 / 20.7 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Blood Group Antigen Binding Adhesin Baba of Helicobacter Pylori Strain 17875 in Complex with Nanobody Nb-ER19
(pdb code 5f7k). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Blood Group Antigen Binding Adhesin Baba of Helicobacter Pylori Strain 17875 in Complex with Nanobody Nb-ER19, PDB code: 5f7k:
In total only one binding site of Sodium was determined in the Blood Group Antigen Binding Adhesin Baba of Helicobacter Pylori Strain 17875 in Complex with Nanobody Nb-ER19, PDB code: 5f7k:
Sodium binding site 1 out of 1 in 5f7k
Go back to
Sodium binding site 1 out
of 1 in the Blood Group Antigen Binding Adhesin Baba of Helicobacter Pylori Strain 17875 in Complex with Nanobody Nb-ER19
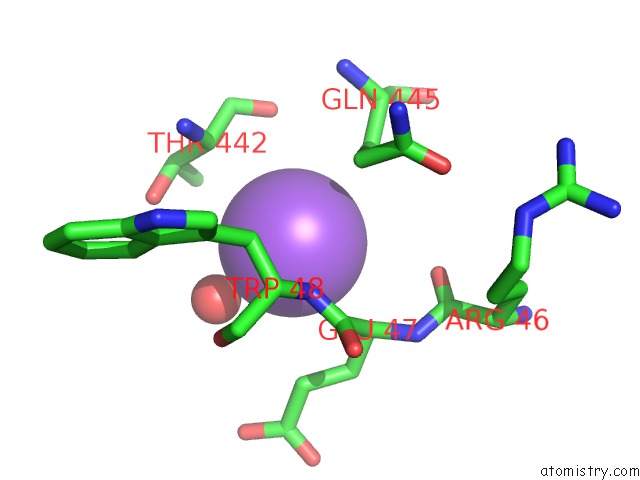
Mono view
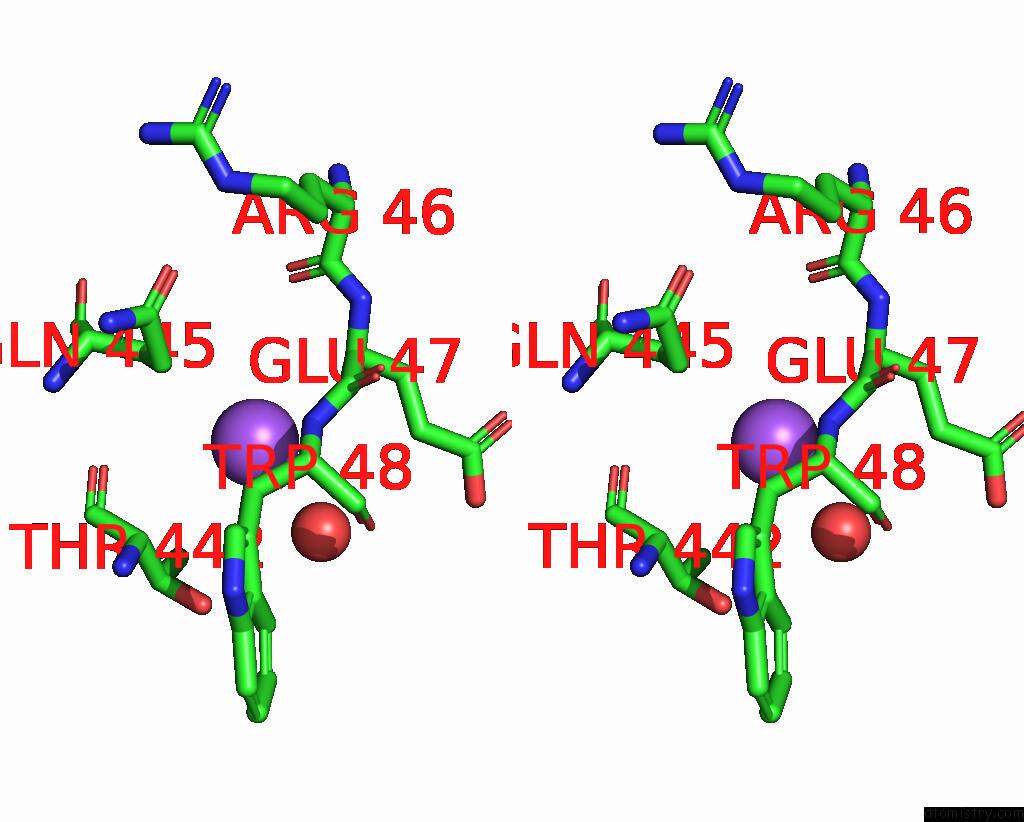
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Blood Group Antigen Binding Adhesin Baba of Helicobacter Pylori Strain 17875 in Complex with Nanobody Nb-ER19 within 5.0Å range:
|
Reference:
K.Moonens,
P.Gideonsson,
S.Subedi,
J.Bugaytsova,
E.Romao,
M.Mendez,
J.Norden,
M.Fallah,
L.Rakhimova,
A.Shevtsova,
M.Lahmann,
G.Castaldo,
K.Brannstrom,
F.Coppens,
A.W.Lo,
T.Ny,
J.V.Solnick,
G.Vandenbussche,
S.Oscarson,
L.Hammarstrom,
A.Arnqvist,
D.E.Berg,
S.Muyldermans,
T.Boren,
H.Remaut.
Structural Insights Into Polymorphic Abo Glycan Binding By Helicobacter Pylori. Cell Host Microbe V. 19 55 2016.
ISSN: ESSN 1934-6069
PubMed: 26764597
DOI: 10.1016/J.CHOM.2015.12.004
Page generated: Sun Aug 17 23:45:46 2025
ISSN: ESSN 1934-6069
PubMed: 26764597
DOI: 10.1016/J.CHOM.2015.12.004
Last articles
Ni in 4AUYNi in 4AUU
Ni in 4ASG
Ni in 4ASF
Ni in 4AL2
Ni in 4AM8
Ni in 4AC7
Ni in 4AI9
Ni in 473D
Ni in 4A8T