Sodium »
PDB 5ddq-5dy9 »
5dgr »
Sodium in PDB 5dgr: Crystal Structure of GH9 Exo-Beta-D-Glucosaminidase PBPRA0520, Glucosamine Complex
Protein crystallography data
The structure of Crystal Structure of GH9 Exo-Beta-D-Glucosaminidase PBPRA0520, Glucosamine Complex, PDB code: 5dgr
was solved by
K.Suzuki,
Y.Honda,
S.Fushinobu,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 28.90 / 1.90 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 62.294, 102.590, 89.804, 90.00, 97.22, 90.00 |
| R / Rfree (%) | 16.2 / 20.4 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Crystal Structure of GH9 Exo-Beta-D-Glucosaminidase PBPRA0520, Glucosamine Complex
(pdb code 5dgr). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 2 binding sites of Sodium where determined in the Crystal Structure of GH9 Exo-Beta-D-Glucosaminidase PBPRA0520, Glucosamine Complex, PDB code: 5dgr:
Jump to Sodium binding site number: 1; 2;
In total 2 binding sites of Sodium where determined in the Crystal Structure of GH9 Exo-Beta-D-Glucosaminidase PBPRA0520, Glucosamine Complex, PDB code: 5dgr:
Jump to Sodium binding site number: 1; 2;
Sodium binding site 1 out of 2 in 5dgr
Go back to
Sodium binding site 1 out
of 2 in the Crystal Structure of GH9 Exo-Beta-D-Glucosaminidase PBPRA0520, Glucosamine Complex

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Crystal Structure of GH9 Exo-Beta-D-Glucosaminidase PBPRA0520, Glucosamine Complex within 5.0Å range:
|
Sodium binding site 2 out of 2 in 5dgr
Go back to
Sodium binding site 2 out
of 2 in the Crystal Structure of GH9 Exo-Beta-D-Glucosaminidase PBPRA0520, Glucosamine Complex

Mono view
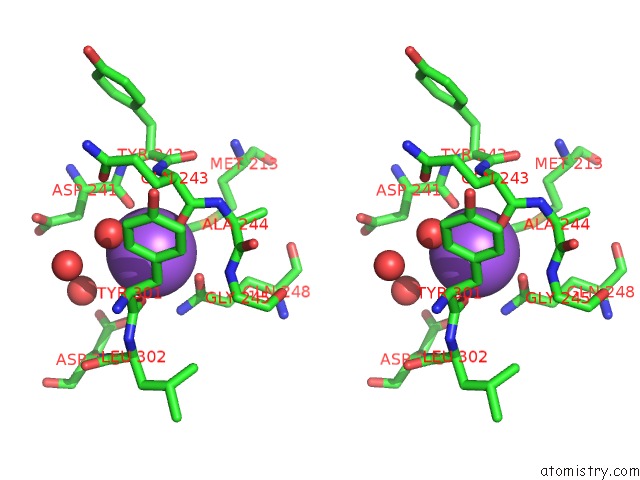
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Crystal Structure of GH9 Exo-Beta-D-Glucosaminidase PBPRA0520, Glucosamine Complex within 5.0Å range:
|
Reference:
Y.Honda,
S.Arai,
K.Suzuki,
M.Kitaoka,
S.Fushinobu.
The Crystal Structure of An Inverting Glycoside Hydrolase Family 9 Exo-Beta-D-Glucosaminidase and the Design of Glycosynthase. Biochem.J. V. 473 463 2016.
ISSN: ESSN 1470-8728
PubMed: 26621872
DOI: 10.1042/BJ20150966
Page generated: Mon Oct 7 20:36:24 2024
ISSN: ESSN 1470-8728
PubMed: 26621872
DOI: 10.1042/BJ20150966
Last articles
Mg in 364DMg in 3A0T
Mg in 357D
Mg in 3A06
Mg in 392D
Mg in 389D
Mg in 388D
Mg in 384D
Mg in 354D
Mg in 383D