Sodium »
PDB 4wxs-4xdu »
4x0s »
Sodium in PDB 4x0s: Hexamer Formed By A Macrocycle Containing A Sequence From Beta-2- Microglobulin (63-69).
Protein crystallography data
The structure of Hexamer Formed By A Macrocycle Containing A Sequence From Beta-2- Microglobulin (63-69)., PDB code: 4x0s
was solved by
R.K.Spencer,
P.J.Salveson,
J.S.Nowick,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 25.62 / 2.03 |
| Space group | H 3 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 51.065, 51.065, 31.423, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 16.5 / 20.2 |
Other elements in 4x0s:
The structure of Hexamer Formed By A Macrocycle Containing A Sequence From Beta-2- Microglobulin (63-69). also contains other interesting chemical elements:
| Iodine | (I) | 1 atom |
| Chlorine | (Cl) | 1 atom |
Sodium Binding Sites:
The binding sites of Sodium atom in the Hexamer Formed By A Macrocycle Containing A Sequence From Beta-2- Microglobulin (63-69).
(pdb code 4x0s). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Hexamer Formed By A Macrocycle Containing A Sequence From Beta-2- Microglobulin (63-69)., PDB code: 4x0s:
In total only one binding site of Sodium was determined in the Hexamer Formed By A Macrocycle Containing A Sequence From Beta-2- Microglobulin (63-69)., PDB code: 4x0s:
Sodium binding site 1 out of 1 in 4x0s
Go back to
Sodium binding site 1 out
of 1 in the Hexamer Formed By A Macrocycle Containing A Sequence From Beta-2- Microglobulin (63-69).
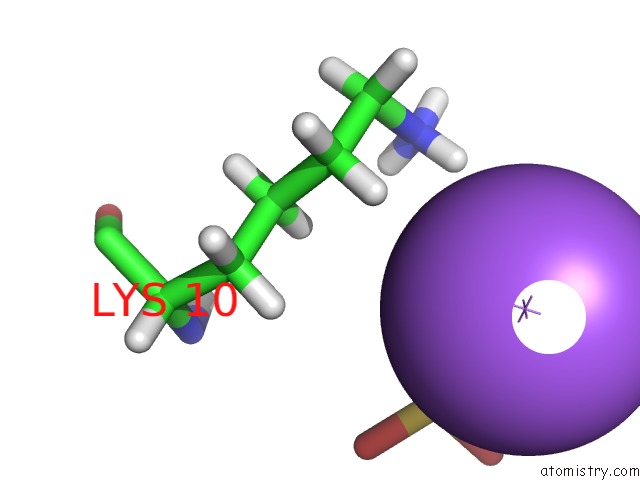
Mono view
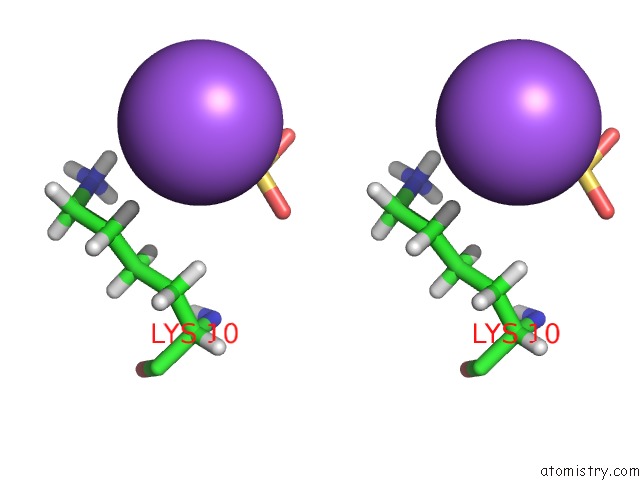
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Hexamer Formed By A Macrocycle Containing A Sequence From Beta-2- Microglobulin (63-69). within 5.0Å range:
|
Reference:
R.K.Spencer,
A.G.Kreutzer,
P.J.Salveson,
H.Li,
J.S.Nowick.
X-Ray Crystallographic Structures of Oligomers of Peptides Derived From Beta 2-Microglobulin. J.Am.Chem.Soc. 2015.
ISSN: ESSN 1520-5126
PubMed: 25915729
DOI: 10.1021/JACS.5B01673
Page generated: Mon Oct 7 18:57:06 2024
ISSN: ESSN 1520-5126
PubMed: 25915729
DOI: 10.1021/JACS.5B01673
Last articles
K in 1JFVK in 1JF8
K in 1JDR
K in 1JCI
K in 1JBS
K in 1JBT
K in 1J95
K in 1JBR
K in 1J5Y
K in 1IS8