Sodium »
PDB 3ma9-3mo9 »
3mj6 »
Sodium in PDB 3mj6: Crystal Structure of the Gammadelta T Cell Costimulatory Receptor Junctional Adhesion Molecule-Like Protein, Jaml
Protein crystallography data
The structure of Crystal Structure of the Gammadelta T Cell Costimulatory Receptor Junctional Adhesion Molecule-Like Protein, Jaml, PDB code: 3mj6
was solved by
P.Verdino,
I.A.Wilson,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.19 |
| Space group | P 43 |
| Cell size a, b, c (Å), α, β, γ (°) | 61.946, 61.946, 82.477, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17 / 21.3 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Crystal Structure of the Gammadelta T Cell Costimulatory Receptor Junctional Adhesion Molecule-Like Protein, Jaml
(pdb code 3mj6). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Crystal Structure of the Gammadelta T Cell Costimulatory Receptor Junctional Adhesion Molecule-Like Protein, Jaml, PDB code: 3mj6:
In total only one binding site of Sodium was determined in the Crystal Structure of the Gammadelta T Cell Costimulatory Receptor Junctional Adhesion Molecule-Like Protein, Jaml, PDB code: 3mj6:
Sodium binding site 1 out of 1 in 3mj6
Go back to
Sodium binding site 1 out
of 1 in the Crystal Structure of the Gammadelta T Cell Costimulatory Receptor Junctional Adhesion Molecule-Like Protein, Jaml
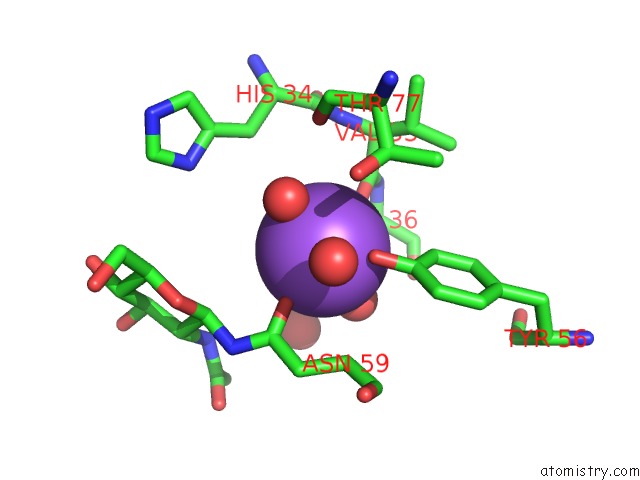
Mono view
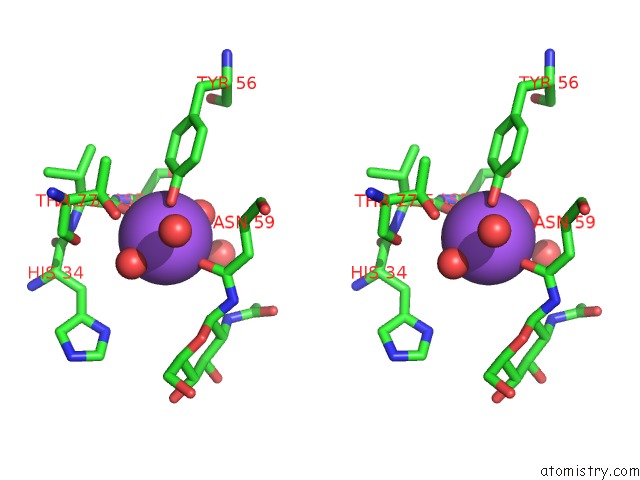
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Crystal Structure of the Gammadelta T Cell Costimulatory Receptor Junctional Adhesion Molecule-Like Protein, Jaml within 5.0Å range:
|
Reference:
P.Verdino,
D.A.Witherden,
W.L.Havran,
I.A.Wilson.
The Molecular Interaction of Car and Jaml Recruits the Central Cell Signal Transducer PI3K. Science V. 329 1210 2010.
ISSN: ISSN 0036-8075
PubMed: 20813955
DOI: 10.1126/SCIENCE.1187996
Page generated: Mon Oct 7 11:31:02 2024
ISSN: ISSN 0036-8075
PubMed: 20813955
DOI: 10.1126/SCIENCE.1187996
Last articles
Ir in 4XW7Ir in 4P5J
Ir in 4P9R
Ir in 4FRG
Ir in 4QKA
Ir in 4NHT
Ir in 4OKA
Ir in 4GXY
Ir in 4NIJ
Ir in 4NHS