Sodium »
PDB 3hwe-3ic9 »
3i4q »
Sodium in PDB 3i4q: Structure of A Putative Inorganic Pyrophosphatase From the Oil- Degrading Bacterium Oleispira Antarctica
Enzymatic activity of Structure of A Putative Inorganic Pyrophosphatase From the Oil- Degrading Bacterium Oleispira Antarctica
All present enzymatic activity of Structure of A Putative Inorganic Pyrophosphatase From the Oil- Degrading Bacterium Oleispira Antarctica:
3.6.1.1;
3.6.1.1;
Protein crystallography data
The structure of Structure of A Putative Inorganic Pyrophosphatase From the Oil- Degrading Bacterium Oleispira Antarctica, PDB code: 3i4q
was solved by
A.U.Singer,
E.Evdokimova,
O.Kagan,
A.M.Edwards,
A.Joachimiak,
A.Savchenko,
Midwest Center For Structural Genomics (Mcsg),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 32.56 / 1.63 |
| Space group | H 3 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 110.666, 110.666, 74.324, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 17.4 / 21.2 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Structure of A Putative Inorganic Pyrophosphatase From the Oil- Degrading Bacterium Oleispira Antarctica
(pdb code 3i4q). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 2 binding sites of Sodium where determined in the Structure of A Putative Inorganic Pyrophosphatase From the Oil- Degrading Bacterium Oleispira Antarctica, PDB code: 3i4q:
Jump to Sodium binding site number: 1; 2;
In total 2 binding sites of Sodium where determined in the Structure of A Putative Inorganic Pyrophosphatase From the Oil- Degrading Bacterium Oleispira Antarctica, PDB code: 3i4q:
Jump to Sodium binding site number: 1; 2;
Sodium binding site 1 out of 2 in 3i4q
Go back to
Sodium binding site 1 out
of 2 in the Structure of A Putative Inorganic Pyrophosphatase From the Oil- Degrading Bacterium Oleispira Antarctica
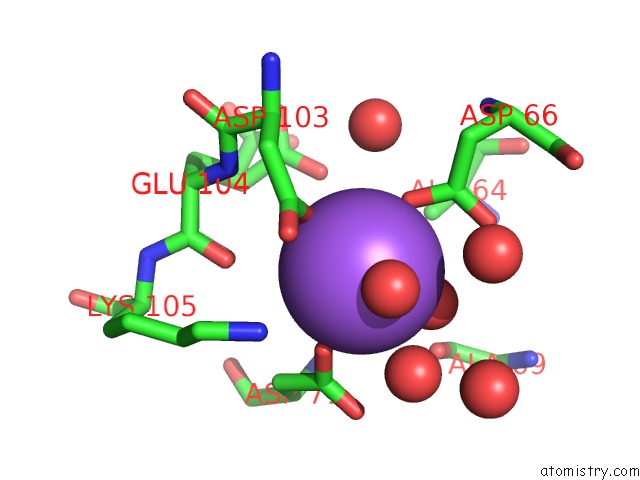
Mono view
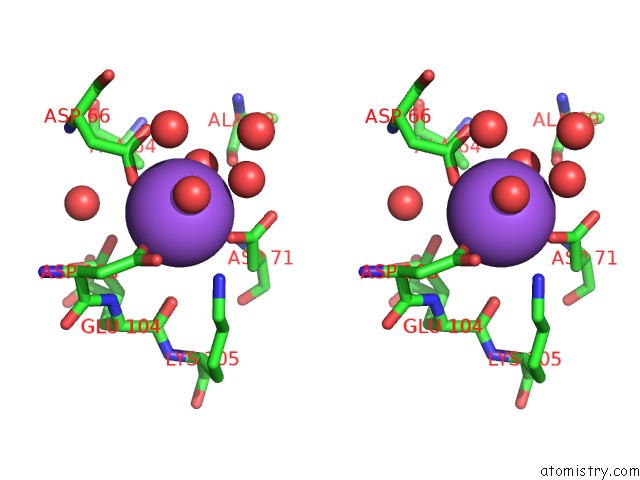
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Structure of A Putative Inorganic Pyrophosphatase From the Oil- Degrading Bacterium Oleispira Antarctica within 5.0Å range:
|
Sodium binding site 2 out of 2 in 3i4q
Go back to
Sodium binding site 2 out
of 2 in the Structure of A Putative Inorganic Pyrophosphatase From the Oil- Degrading Bacterium Oleispira Antarctica
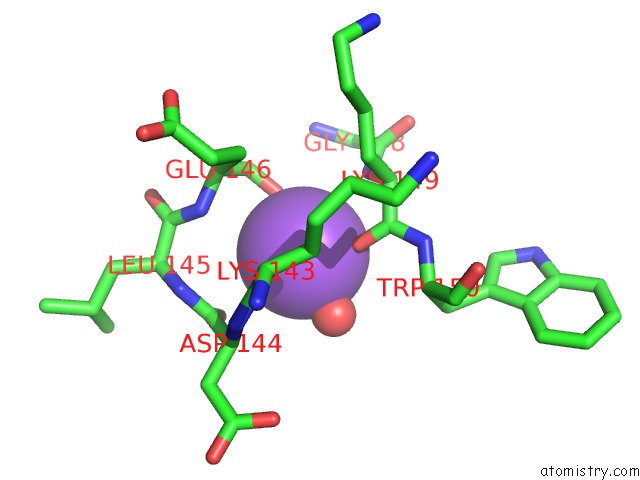
Mono view
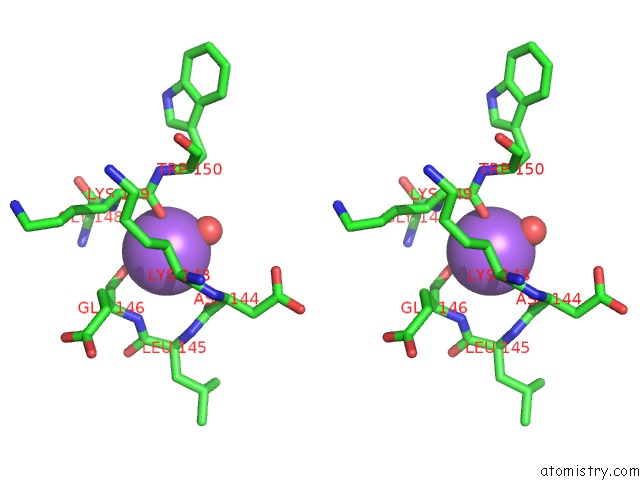
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Structure of A Putative Inorganic Pyrophosphatase From the Oil- Degrading Bacterium Oleispira Antarctica within 5.0Å range:
|
Reference:
M.Kube,
T.N.Chernikova,
Y.Al-Ramahi,
A.Beloqui,
N.Lopez-Cortez,
M.E.Guazzaroni,
H.J.Heipieper,
S.Klages,
O.R.Kotsyurbenko,
I.Langer,
T.Y.Nechitaylo,
H.Lunsdorf,
M.Fernandez,
S.Juarez,
S.Ciordia,
A.Singer,
O.Kagan,
O.Egorova,
P.Alain Petit,
P.Stogios,
Y.Kim,
A.Tchigvintsev,
R.Flick,
R.Denaro,
M.Genovese,
J.P.Albar,
O.N.Reva,
M.Martinez-Gomariz,
H.Tran,
M.Ferrer,
A.Savchenko,
A.F.Yakunin,
M.M.Yakimov,
O.V.Golyshina,
R.Reinhardt,
P.N.Golyshin.
Genome Sequence and Functional Genomic Analysis of the Oil-Degrading Bacterium Oleispira Antarctica. Nat Commun V. 4 2156 2013.
ISSN: ESSN 2041-1723
PubMed: 23877221
DOI: 10.1038/NCOMMS3156
Page generated: Mon Oct 7 10:34:59 2024
ISSN: ESSN 2041-1723
PubMed: 23877221
DOI: 10.1038/NCOMMS3156
Last articles
K in 7ACCK in 7ADI
K in 7ABN
K in 6ZRW
K in 7A0S
K in 7A56
K in 6ZXZ
K in 7A4V
K in 6ZW1
K in 6ZT3