Sodium »
PDB 3gdx-3h09 »
3geg »
Sodium in PDB 3geg: Fingerprint and Structural Analysis of A Scor Enzyme with Its Bound Cofactor From Clostridium Thermocellum
Protein crystallography data
The structure of Fingerprint and Structural Analysis of A Scor Enzyme with Its Bound Cofactor From Clostridium Thermocellum, PDB code: 3geg
was solved by
R.Huether,
Z.J.Liu,
H.Xu,
B.C.Wang,
V.Pletnev,
Q.Mao,
T.Umland,
W.Duax,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 43.94 / 2.10 |
| Space group | I 41 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 124.272, 124.272, 161.208, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.7 / 20 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Fingerprint and Structural Analysis of A Scor Enzyme with Its Bound Cofactor From Clostridium Thermocellum
(pdb code 3geg). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 2 binding sites of Sodium where determined in the Fingerprint and Structural Analysis of A Scor Enzyme with Its Bound Cofactor From Clostridium Thermocellum, PDB code: 3geg:
Jump to Sodium binding site number: 1; 2;
In total 2 binding sites of Sodium where determined in the Fingerprint and Structural Analysis of A Scor Enzyme with Its Bound Cofactor From Clostridium Thermocellum, PDB code: 3geg:
Jump to Sodium binding site number: 1; 2;
Sodium binding site 1 out of 2 in 3geg
Go back to
Sodium binding site 1 out
of 2 in the Fingerprint and Structural Analysis of A Scor Enzyme with Its Bound Cofactor From Clostridium Thermocellum

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Fingerprint and Structural Analysis of A Scor Enzyme with Its Bound Cofactor From Clostridium Thermocellum within 5.0Å range:
|
Sodium binding site 2 out of 2 in 3geg
Go back to
Sodium binding site 2 out
of 2 in the Fingerprint and Structural Analysis of A Scor Enzyme with Its Bound Cofactor From Clostridium Thermocellum
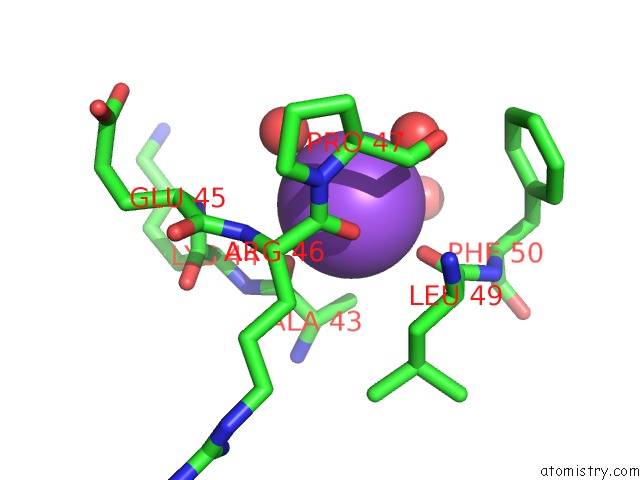
Mono view
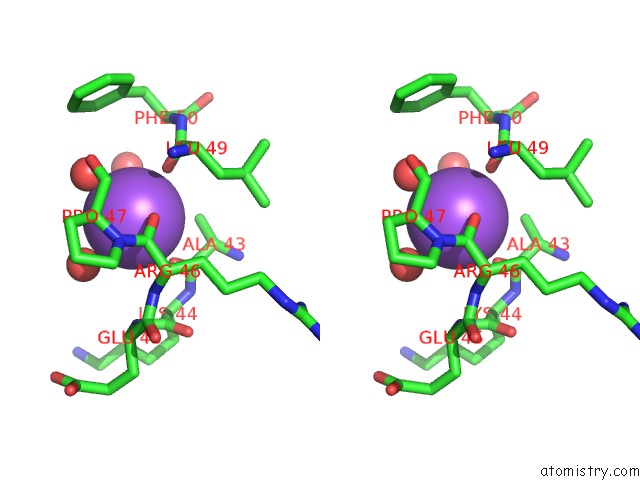
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Fingerprint and Structural Analysis of A Scor Enzyme with Its Bound Cofactor From Clostridium Thermocellum within 5.0Å range:
|
Reference:
R.Huether,
Z.J.Liu,
H.Xu,
B.C.Wang,
V.Z.Pletnev,
Q.Mao,
W.L.Duax,
T.C.Umland.
Sequence Fingerprint and Structural Analysis of the Scor Enzyme A3DFK9 From Clostridium Thermocellum. Proteins V. 78 603 2010.
ISSN: ISSN 0887-3585
PubMed: 19774618
DOI: 10.1002/PROT.22584
Page generated: Mon Oct 7 10:15:08 2024
ISSN: ISSN 0887-3585
PubMed: 19774618
DOI: 10.1002/PROT.22584
Last articles
Mg in 4WFNMg in 4WF9
Mg in 4WFB
Mg in 4WFM
Mg in 4WFL
Mg in 4WCE
Mg in 4WF7
Mg in 4WEC
Mg in 4WEO
Mg in 4WCW