Sodium »
PDB 3bjp-3c7h »
3c52 »
Sodium in PDB 3c52: Class II Fructose-1,6-Bisphosphate Aldolase From Helicobacter Pylori in Complex with Phosphoglycolohydroxamic Acid, A Competitive Inhibitor
Enzymatic activity of Class II Fructose-1,6-Bisphosphate Aldolase From Helicobacter Pylori in Complex with Phosphoglycolohydroxamic Acid, A Competitive Inhibitor
All present enzymatic activity of Class II Fructose-1,6-Bisphosphate Aldolase From Helicobacter Pylori in Complex with Phosphoglycolohydroxamic Acid, A Competitive Inhibitor:
4.1.2.13;
4.1.2.13;
Protein crystallography data
The structure of Class II Fructose-1,6-Bisphosphate Aldolase From Helicobacter Pylori in Complex with Phosphoglycolohydroxamic Acid, A Competitive Inhibitor, PDB code: 3c52
was solved by
M.Coincon,
J.Sygusch,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 32.00 / 2.30 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 39.242, 85.305, 90.818, 90.00, 100.16, 90.00 |
| R / Rfree (%) | 20.4 / 24.9 |
Other elements in 3c52:
The structure of Class II Fructose-1,6-Bisphosphate Aldolase From Helicobacter Pylori in Complex with Phosphoglycolohydroxamic Acid, A Competitive Inhibitor also contains other interesting chemical elements:
| Zinc | (Zn) | 2 atoms |
| Calcium | (Ca) | 2 atoms |
Sodium Binding Sites:
The binding sites of Sodium atom in the Class II Fructose-1,6-Bisphosphate Aldolase From Helicobacter Pylori in Complex with Phosphoglycolohydroxamic Acid, A Competitive Inhibitor
(pdb code 3c52). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 2 binding sites of Sodium where determined in the Class II Fructose-1,6-Bisphosphate Aldolase From Helicobacter Pylori in Complex with Phosphoglycolohydroxamic Acid, A Competitive Inhibitor, PDB code: 3c52:
Jump to Sodium binding site number: 1; 2;
In total 2 binding sites of Sodium where determined in the Class II Fructose-1,6-Bisphosphate Aldolase From Helicobacter Pylori in Complex with Phosphoglycolohydroxamic Acid, A Competitive Inhibitor, PDB code: 3c52:
Jump to Sodium binding site number: 1; 2;
Sodium binding site 1 out of 2 in 3c52
Go back to
Sodium binding site 1 out
of 2 in the Class II Fructose-1,6-Bisphosphate Aldolase From Helicobacter Pylori in Complex with Phosphoglycolohydroxamic Acid, A Competitive Inhibitor
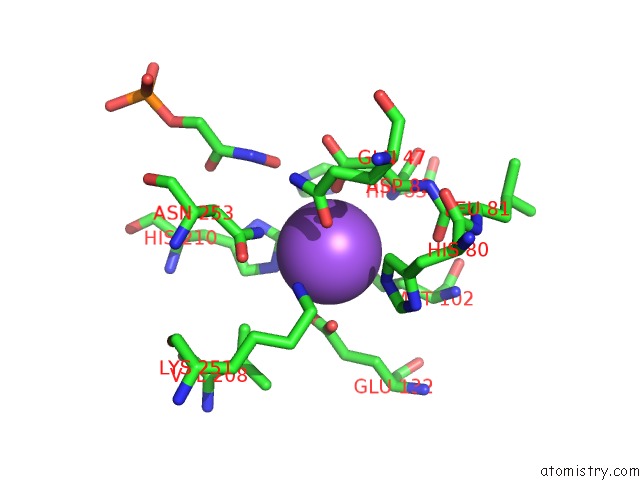
Mono view
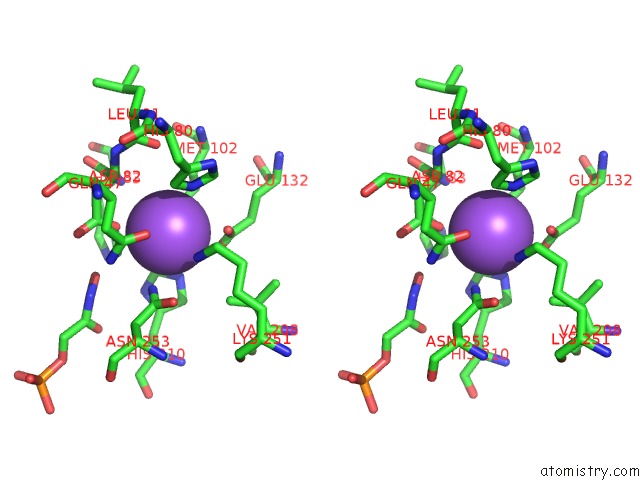
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Class II Fructose-1,6-Bisphosphate Aldolase From Helicobacter Pylori in Complex with Phosphoglycolohydroxamic Acid, A Competitive Inhibitor within 5.0Å range:
|
Sodium binding site 2 out of 2 in 3c52
Go back to
Sodium binding site 2 out
of 2 in the Class II Fructose-1,6-Bisphosphate Aldolase From Helicobacter Pylori in Complex with Phosphoglycolohydroxamic Acid, A Competitive Inhibitor
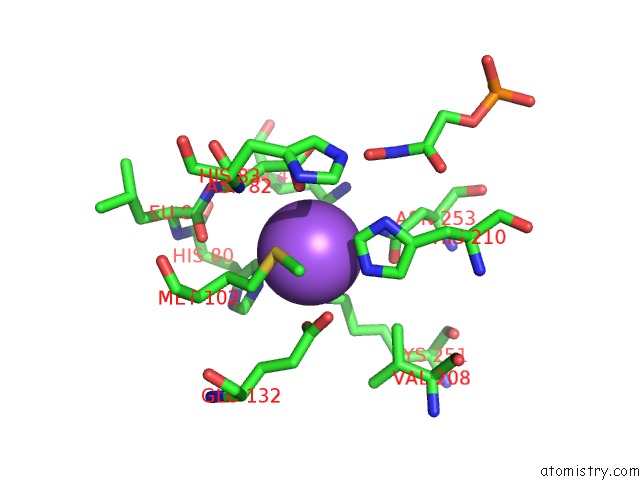
Mono view
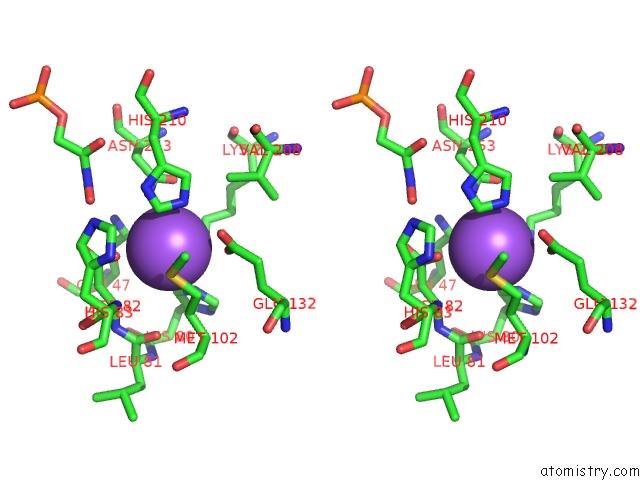
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Class II Fructose-1,6-Bisphosphate Aldolase From Helicobacter Pylori in Complex with Phosphoglycolohydroxamic Acid, A Competitive Inhibitor within 5.0Å range:
|
Reference:
M.Fonvielle,
M.Coincon,
R.Daher,
N.Desbenoit,
K.Kosieradzka,
N.Barilone,
B.Gicquel,
J.Sygusch,
M.Jackson,
M.Therisod.
Synthesis and Biochemical Evaluation of Selective Inhibitors of Class II Fructose Bisphosphate Aldolases: Towards New Synthetic Antibiotics. Chemistry V. 14 8521 2008.
ISSN: ISSN 0947-6539
PubMed: 18688832
DOI: 10.1002/CHEM.200800857
Page generated: Mon Oct 7 06:09:22 2024
ISSN: ISSN 0947-6539
PubMed: 18688832
DOI: 10.1002/CHEM.200800857
Last articles
K in 7ZEBK in 7ZDP
K in 7ZDZ
K in 7ZDM
K in 7ZDJ
K in 7Z4D
K in 7ZDH
K in 7ZD6
K in 7ZD4
K in 7ZD3