Sodium »
PDB 1zum-2aoc »
1zv8 »
Sodium in PDB 1zv8: A Structure-Based Mechanism of Sars Virus Membrane Fusion
Protein crystallography data
The structure of A Structure-Based Mechanism of Sars Virus Membrane Fusion, PDB code: 1zv8
was solved by
Y.Deng,
J.Liu,
Q.Zheng,
W.Yong,
J.Dai,
M.Lu,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 70.71 / 1.94 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 31.054, 55.565, 71.678, 81.26, 87.08, 84.10 |
| R / Rfree (%) | 20.3 / 27.4 |
Other elements in 1zv8:
The structure of A Structure-Based Mechanism of Sars Virus Membrane Fusion also contains other interesting chemical elements:
| Arsenic | (As) | 1 atom |
| Zinc | (Zn) | 2 atoms |
Sodium Binding Sites:
The binding sites of Sodium atom in the A Structure-Based Mechanism of Sars Virus Membrane Fusion
(pdb code 1zv8). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 6 binding sites of Sodium where determined in the A Structure-Based Mechanism of Sars Virus Membrane Fusion, PDB code: 1zv8:
Jump to Sodium binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Sodium where determined in the A Structure-Based Mechanism of Sars Virus Membrane Fusion, PDB code: 1zv8:
Jump to Sodium binding site number: 1; 2; 3; 4; 5; 6;
Sodium binding site 1 out of 6 in 1zv8
Go back to
Sodium binding site 1 out
of 6 in the A Structure-Based Mechanism of Sars Virus Membrane Fusion
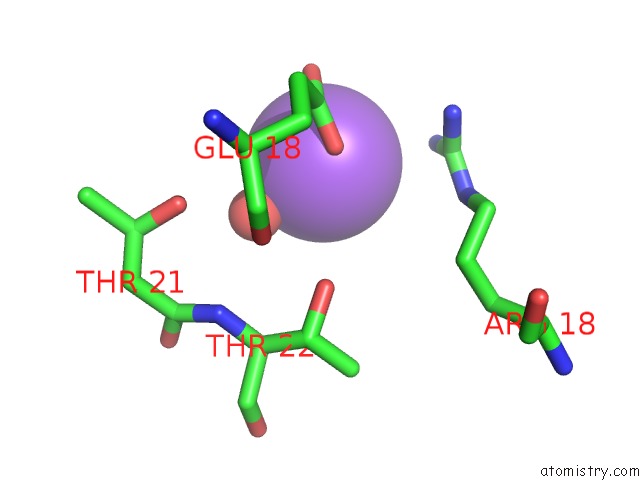
Mono view
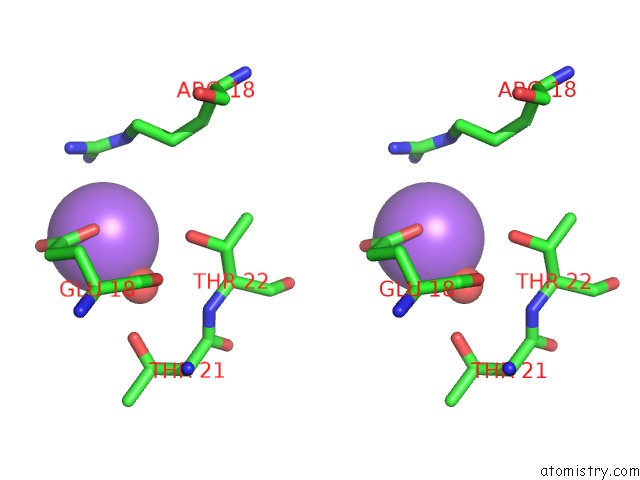
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of A Structure-Based Mechanism of Sars Virus Membrane Fusion within 5.0Å range:
|
Sodium binding site 2 out of 6 in 1zv8
Go back to
Sodium binding site 2 out
of 6 in the A Structure-Based Mechanism of Sars Virus Membrane Fusion
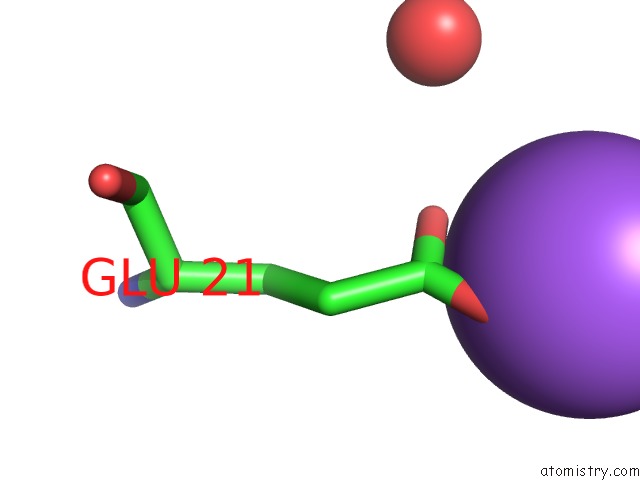
Mono view
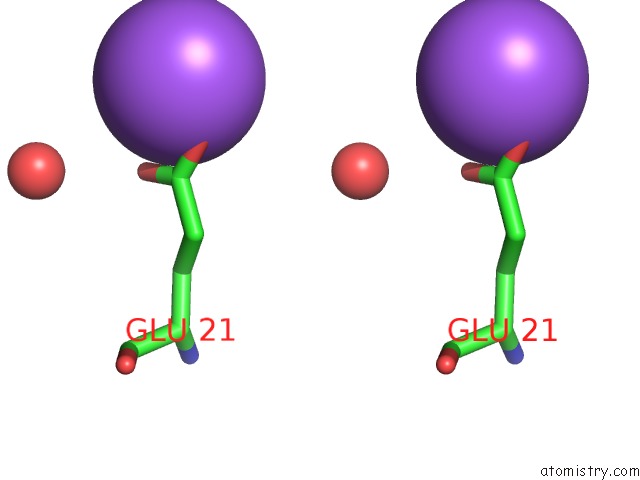
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of A Structure-Based Mechanism of Sars Virus Membrane Fusion within 5.0Å range:
|
Sodium binding site 3 out of 6 in 1zv8
Go back to
Sodium binding site 3 out
of 6 in the A Structure-Based Mechanism of Sars Virus Membrane Fusion
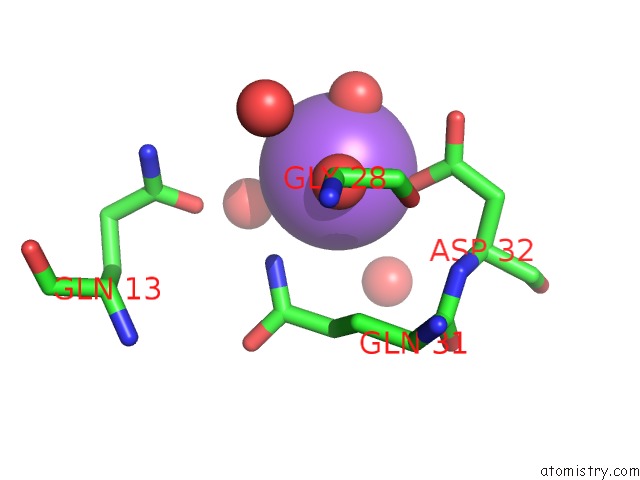
Mono view
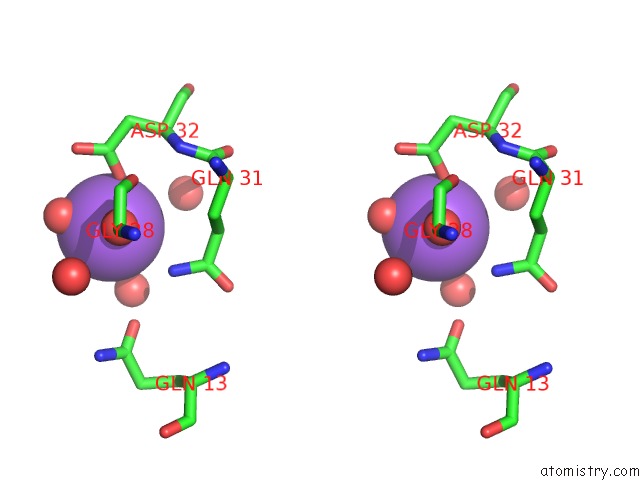
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 3 of A Structure-Based Mechanism of Sars Virus Membrane Fusion within 5.0Å range:
|
Sodium binding site 4 out of 6 in 1zv8
Go back to
Sodium binding site 4 out
of 6 in the A Structure-Based Mechanism of Sars Virus Membrane Fusion
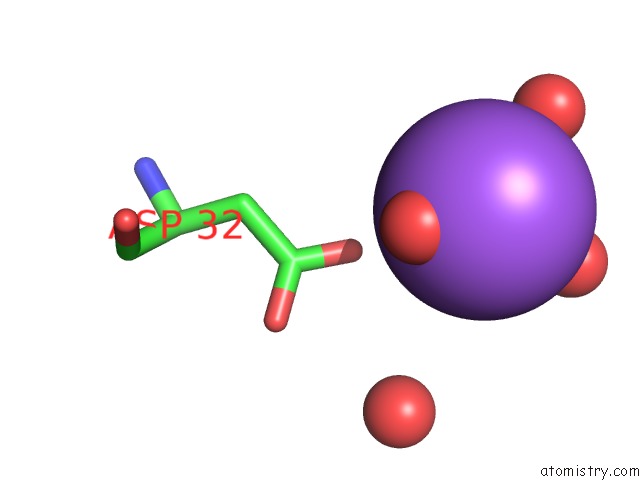
Mono view
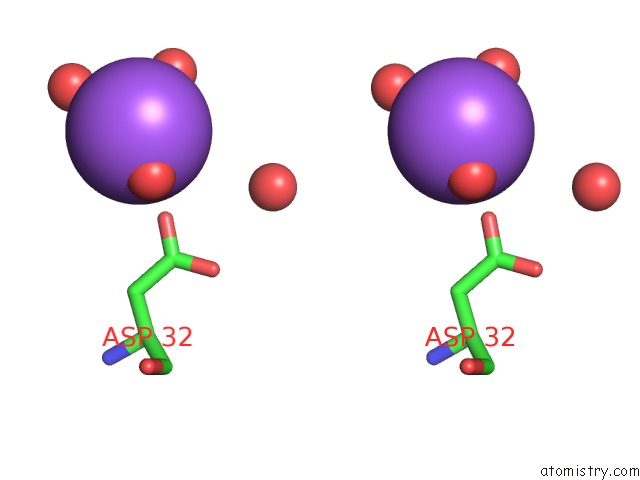
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 4 of A Structure-Based Mechanism of Sars Virus Membrane Fusion within 5.0Å range:
|
Sodium binding site 5 out of 6 in 1zv8
Go back to
Sodium binding site 5 out
of 6 in the A Structure-Based Mechanism of Sars Virus Membrane Fusion
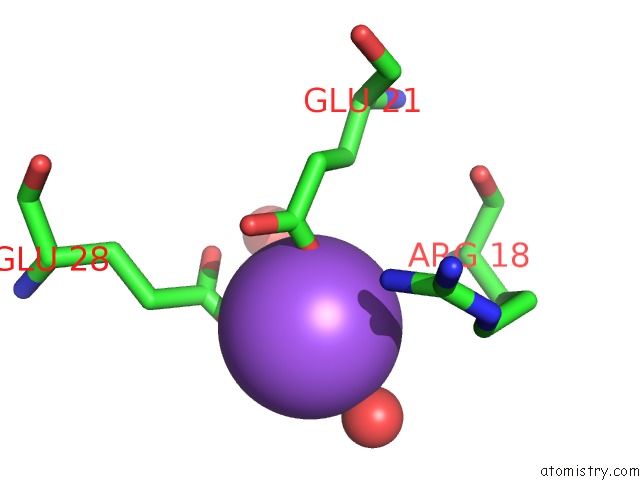
Mono view
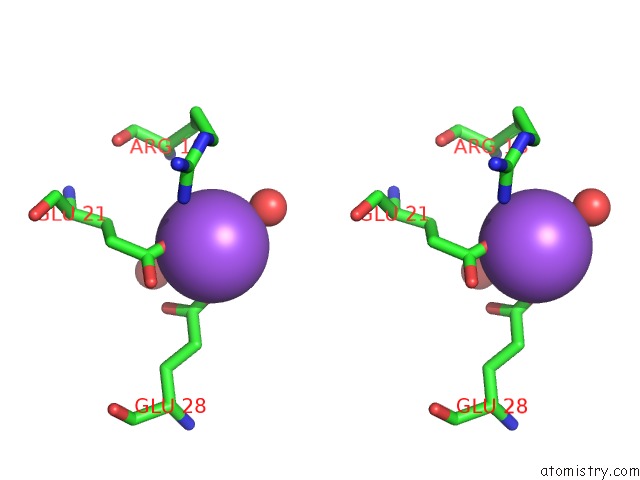
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 5 of A Structure-Based Mechanism of Sars Virus Membrane Fusion within 5.0Å range:
|
Sodium binding site 6 out of 6 in 1zv8
Go back to
Sodium binding site 6 out
of 6 in the A Structure-Based Mechanism of Sars Virus Membrane Fusion
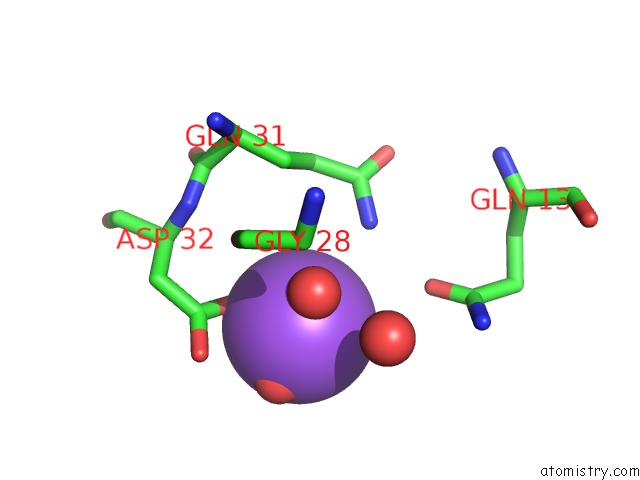
Mono view
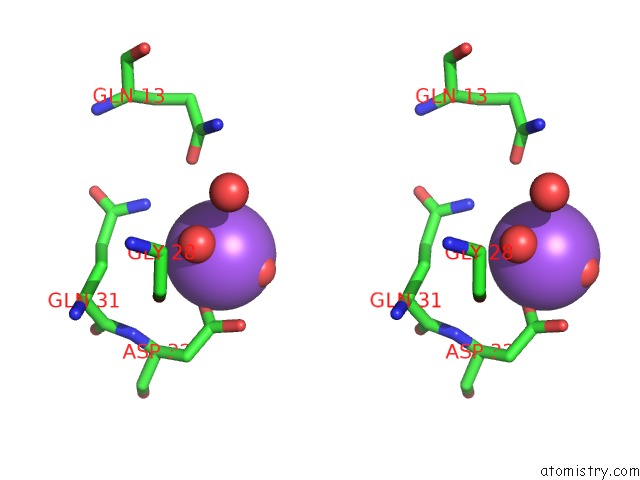
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 6 of A Structure-Based Mechanism of Sars Virus Membrane Fusion within 5.0Å range:
|
Reference:
Y.Deng,
J.Liu,
Q.Zheng,
W.Yong,
M.Lu.
Structures and Polymorphic Interactions of Two Heptad-Repeat Regions of the Sars Virus S2 Protein. Structure V. 14 889 2006.
ISSN: ISSN 0969-2126
PubMed: 16698550
DOI: 10.1016/J.STR.2006.03.007
Page generated: Mon Oct 7 01:45:39 2024
ISSN: ISSN 0969-2126
PubMed: 16698550
DOI: 10.1016/J.STR.2006.03.007
Last articles
I in 6YR6I in 6YRB
I in 6YT2
I in 6YGD
I in 6YGC
I in 6YGA
I in 6YGB
I in 6Y54
I in 6XYB
I in 6XYU