Sodium »
PDB 8oyw-8pkv »
8ph4 »
Sodium in PDB 8ph4: Co-Crystal Structure of the Sars-COV2 Main Protease NSP5 with An Uracil-Carrying X77-Like Inhibitor
Enzymatic activity of Co-Crystal Structure of the Sars-COV2 Main Protease NSP5 with An Uracil-Carrying X77-Like Inhibitor
All present enzymatic activity of Co-Crystal Structure of the Sars-COV2 Main Protease NSP5 with An Uracil-Carrying X77-Like Inhibitor:
3.4.22.69;
3.4.22.69;
Protein crystallography data
The structure of Co-Crystal Structure of the Sars-COV2 Main Protease NSP5 with An Uracil-Carrying X77-Like Inhibitor, PDB code: 8ph4
was solved by
T.Barthel,
N.Altincekic,
N.Jores,
J.Wollenhaupt,
M.S.Weiss,
H.Schwalbe,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 49.28 / 1.69 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 67.69, 100.04, 103.37, 90, 90, 90 |
| R / Rfree (%) | 22.3 / 24.9 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Co-Crystal Structure of the Sars-COV2 Main Protease NSP5 with An Uracil-Carrying X77-Like Inhibitor
(pdb code 8ph4). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 2 binding sites of Sodium where determined in the Co-Crystal Structure of the Sars-COV2 Main Protease NSP5 with An Uracil-Carrying X77-Like Inhibitor, PDB code: 8ph4:
Jump to Sodium binding site number: 1; 2;
In total 2 binding sites of Sodium where determined in the Co-Crystal Structure of the Sars-COV2 Main Protease NSP5 with An Uracil-Carrying X77-Like Inhibitor, PDB code: 8ph4:
Jump to Sodium binding site number: 1; 2;
Sodium binding site 1 out of 2 in 8ph4
Go back to
Sodium binding site 1 out
of 2 in the Co-Crystal Structure of the Sars-COV2 Main Protease NSP5 with An Uracil-Carrying X77-Like Inhibitor
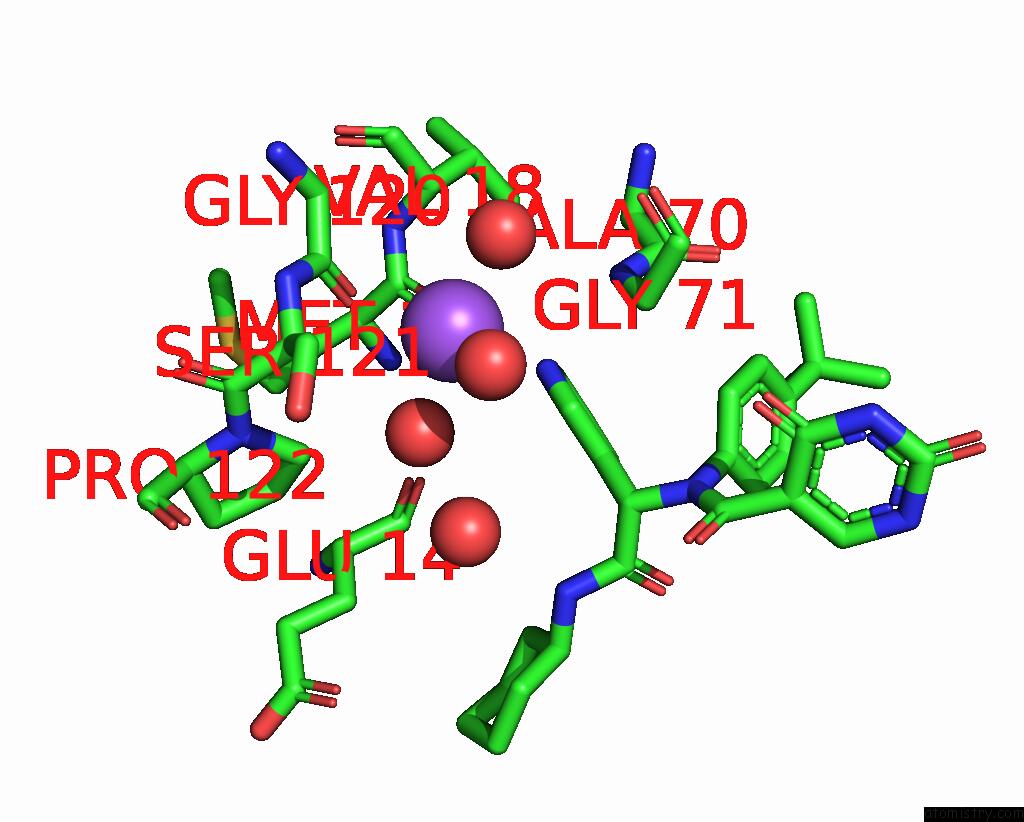
Mono view
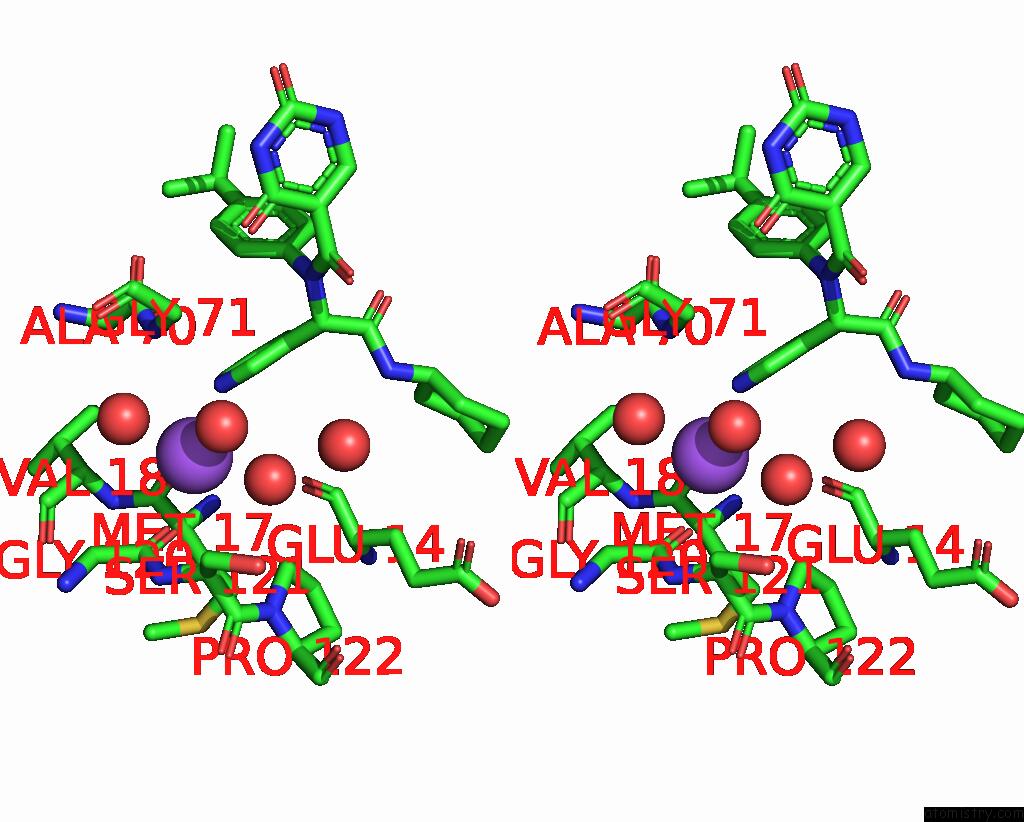
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Co-Crystal Structure of the Sars-COV2 Main Protease NSP5 with An Uracil-Carrying X77-Like Inhibitor within 5.0Å range:
|
Sodium binding site 2 out of 2 in 8ph4
Go back to
Sodium binding site 2 out
of 2 in the Co-Crystal Structure of the Sars-COV2 Main Protease NSP5 with An Uracil-Carrying X77-Like Inhibitor
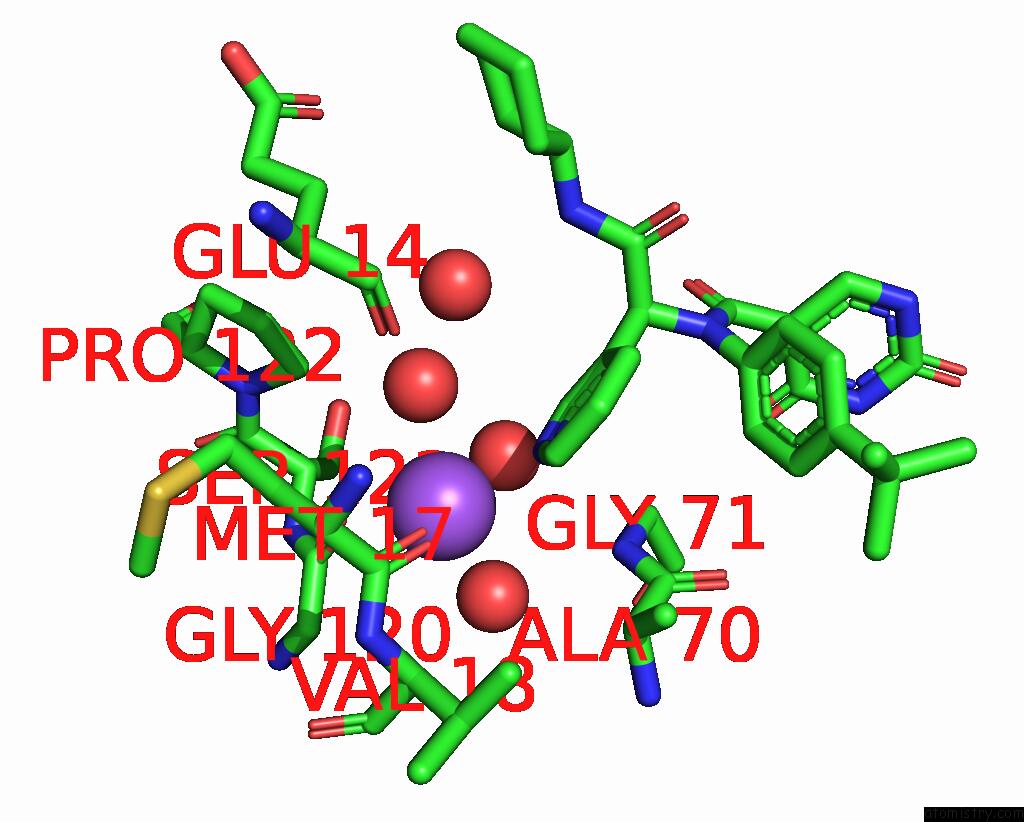
Mono view
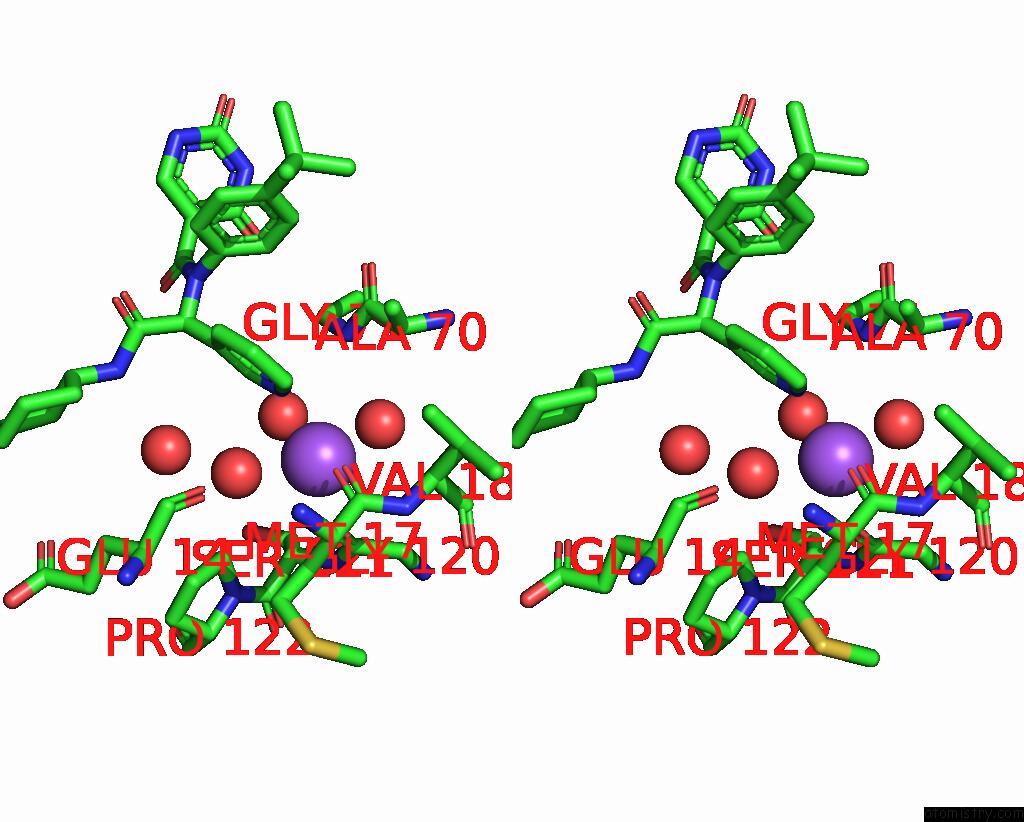
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Co-Crystal Structure of the Sars-COV2 Main Protease NSP5 with An Uracil-Carrying X77-Like Inhibitor within 5.0Å range:
|
Reference:
N.Altincekic,
N.Jores,
F.Lohr,
C.Richter,
C.Ehrhardt,
M.J.J.Blommers,
H.Berg,
S.Ozturk,
S.L.Gande,
V.Linhard,
J.Orts,
M.J.Abi Saad,
M.Butikofer,
J.Kaderli,
B.G.Karlsson,
U.Brath,
M.Hedenstrom,
G.Grobner,
U.H.Sauer,
A.Perrakis,
J.Langer,
L.Banci,
F.Cantini,
M.Fragai,
D.Grifagni,
T.Barthel,
J.Wollenhaupt,
M.S.Weiss,
A.Robertson,
A.Bax,
S.Sreeramulu,
H.Schwalbe.
Targeting the Main Protease (M Pro , NSP5) By Growth of Fragment Scaffolds Exploiting Structure-Based Methodologies. Acs Chem.Biol. 2024.
ISSN: ESSN 1554-8937
PubMed: 38232960
DOI: 10.1021/ACSCHEMBIO.3C00720
Page generated: Mon Aug 18 15:01:14 2025
ISSN: ESSN 1554-8937
PubMed: 38232960
DOI: 10.1021/ACSCHEMBIO.3C00720
Last articles
Ni in 3FMSNi in 3FH3
Ni in 3EYY
Ni in 3FMD
Ni in 3FFO
Ni in 3EGJ
Ni in 3F0A
Ni in 3EXX
Ni in 3ESK
Ni in 3E3U