Sodium in PDB 8fww: Structure of the Ligand-Binding and Transmembrane Domains of Kainate Receptor GLUK2 in Complex with the Positive Allosteric Modulator BPAM344 and Noncompetitive Inhibitor Perampanel
Other elements in 8fww:
The structure of Structure of the Ligand-Binding and Transmembrane Domains of Kainate Receptor GLUK2 in Complex with the Positive Allosteric Modulator BPAM344 and Noncompetitive Inhibitor Perampanel also contains other interesting chemical elements:
| Chlorine | (Cl) | 2 atoms |
| Fluorine | (F) | 4 atoms |
Sodium Binding Sites:
The binding sites of Sodium atom in the Structure of the Ligand-Binding and Transmembrane Domains of Kainate Receptor GLUK2 in Complex with the Positive Allosteric Modulator BPAM344 and Noncompetitive Inhibitor Perampanel
(pdb code 8fww). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 7 binding sites of Sodium where determined in the Structure of the Ligand-Binding and Transmembrane Domains of Kainate Receptor GLUK2 in Complex with the Positive Allosteric Modulator BPAM344 and Noncompetitive Inhibitor Perampanel, PDB code: 8fww:
Jump to Sodium binding site number: 1; 2; 3; 4; 5; 6; 7;
In total 7 binding sites of Sodium where determined in the Structure of the Ligand-Binding and Transmembrane Domains of Kainate Receptor GLUK2 in Complex with the Positive Allosteric Modulator BPAM344 and Noncompetitive Inhibitor Perampanel, PDB code: 8fww:
Jump to Sodium binding site number: 1; 2; 3; 4; 5; 6; 7;
Sodium binding site 1 out of 7 in 8fww
Go back to
Sodium binding site 1 out
of 7 in the Structure of the Ligand-Binding and Transmembrane Domains of Kainate Receptor GLUK2 in Complex with the Positive Allosteric Modulator BPAM344 and Noncompetitive Inhibitor Perampanel
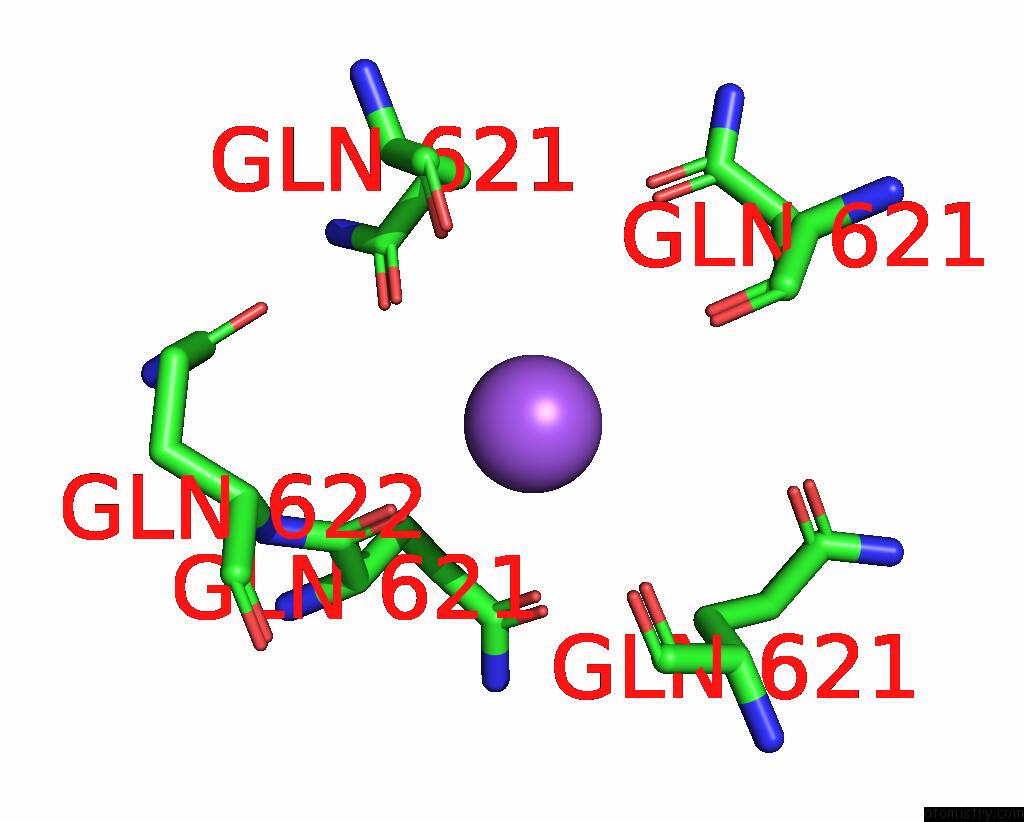
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Structure of the Ligand-Binding and Transmembrane Domains of Kainate Receptor GLUK2 in Complex with the Positive Allosteric Modulator BPAM344 and Noncompetitive Inhibitor Perampanel within 5.0Å range:
|
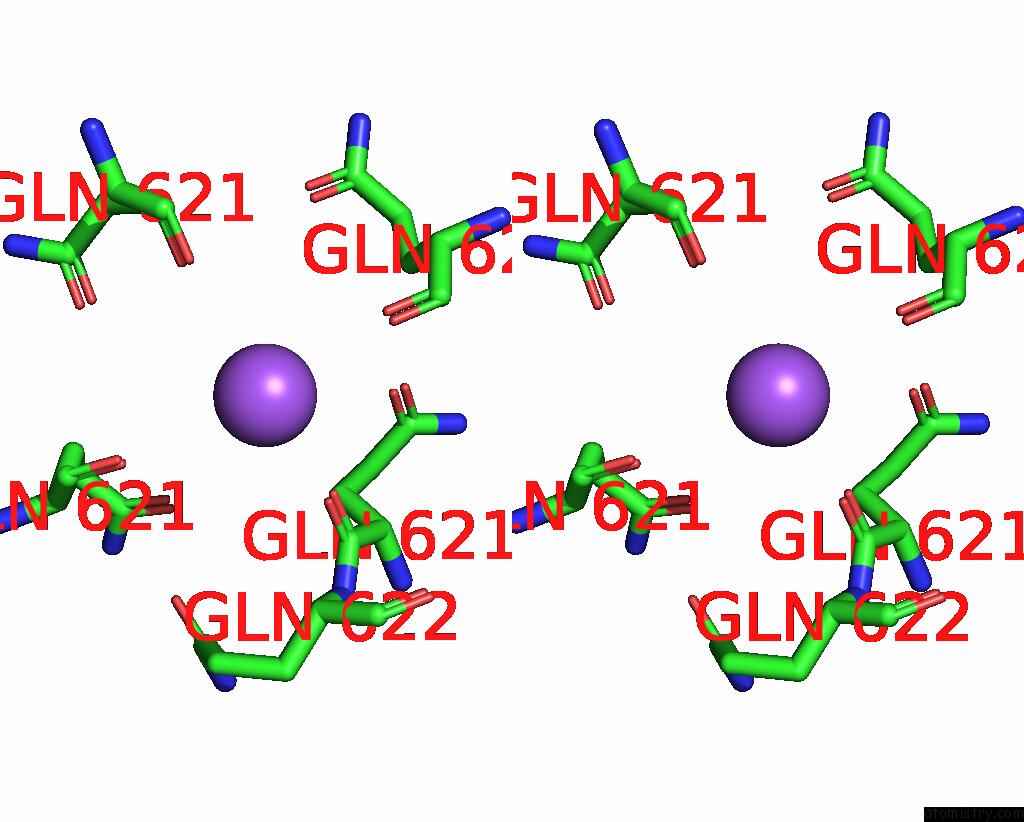
Sodium binding site 2 out of 7 in 8fww
Go back to
Sodium binding site 2 out
of 7 in the Structure of the Ligand-Binding and Transmembrane Domains of Kainate Receptor GLUK2 in Complex with the Positive Allosteric Modulator BPAM344 and Noncompetitive Inhibitor Perampanel

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Structure of the Ligand-Binding and Transmembrane Domains of Kainate Receptor GLUK2 in Complex with the Positive Allosteric Modulator BPAM344 and Noncompetitive Inhibitor Perampanel within 5.0Å range:
|
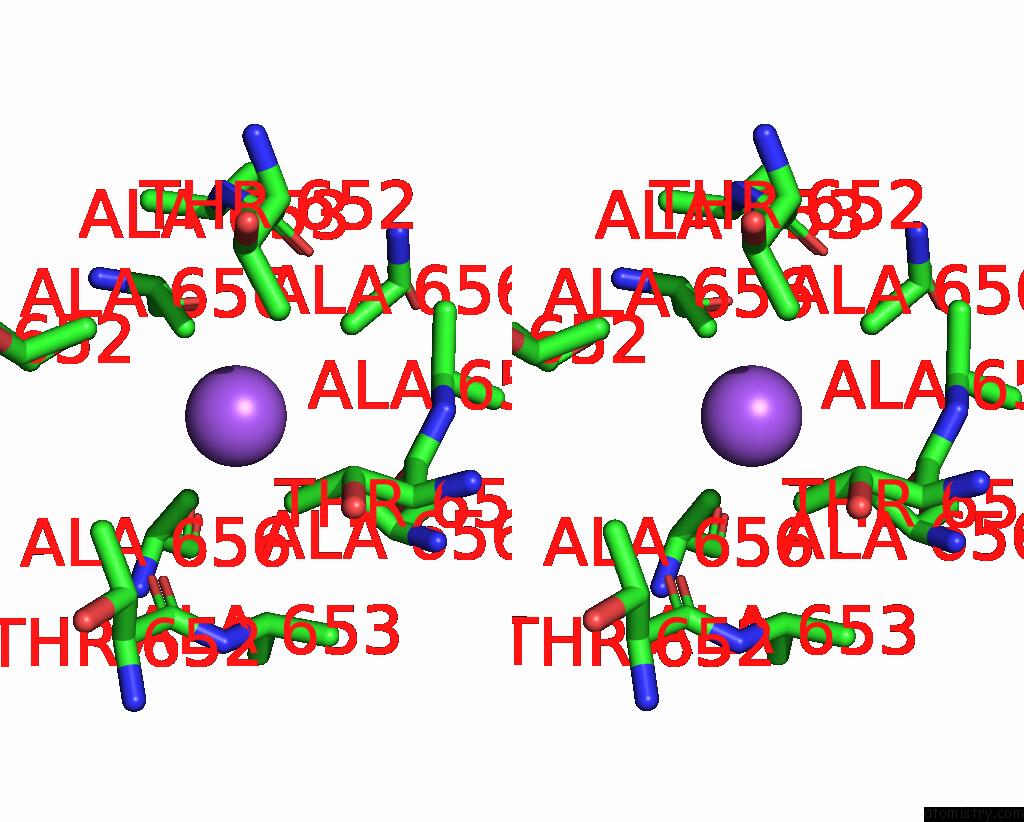
Sodium binding site 3 out of 7 in 8fww
Go back to
Sodium binding site 3 out
of 7 in the Structure of the Ligand-Binding and Transmembrane Domains of Kainate Receptor GLUK2 in Complex with the Positive Allosteric Modulator BPAM344 and Noncompetitive Inhibitor Perampanel

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 3 of Structure of the Ligand-Binding and Transmembrane Domains of Kainate Receptor GLUK2 in Complex with the Positive Allosteric Modulator BPAM344 and Noncompetitive Inhibitor Perampanel within 5.0Å range:
|
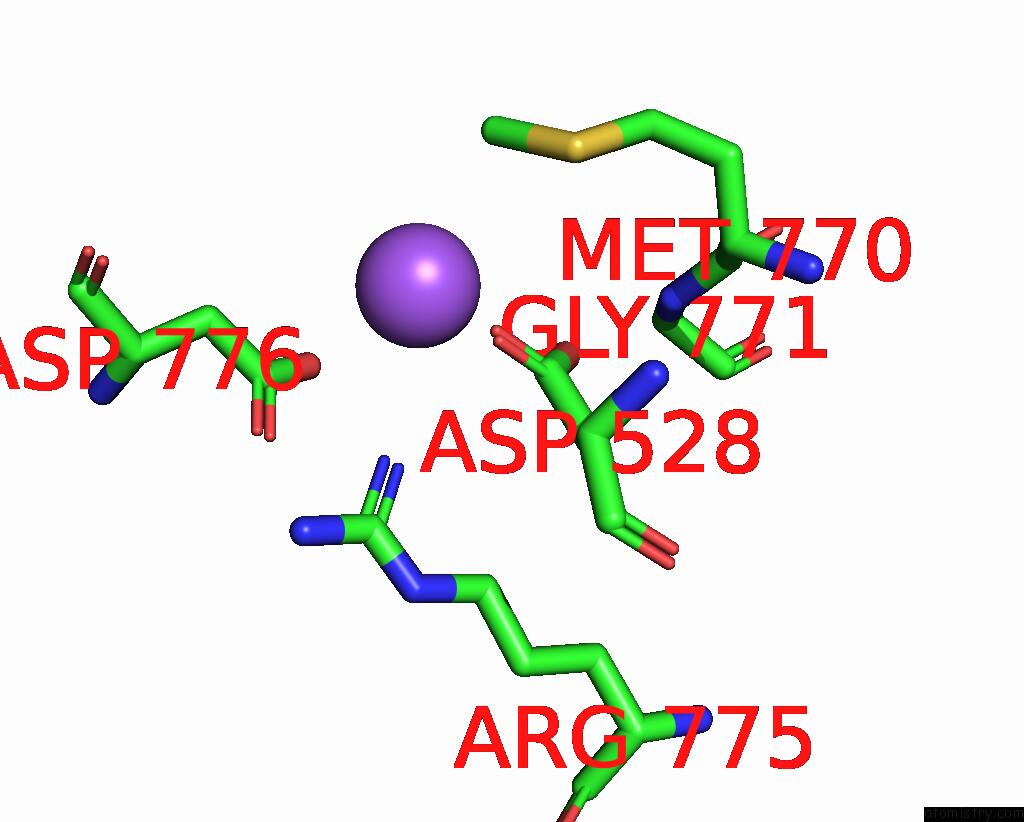
Sodium binding site 4 out of 7 in 8fww
Go back to
Sodium binding site 4 out
of 7 in the Structure of the Ligand-Binding and Transmembrane Domains of Kainate Receptor GLUK2 in Complex with the Positive Allosteric Modulator BPAM344 and Noncompetitive Inhibitor Perampanel
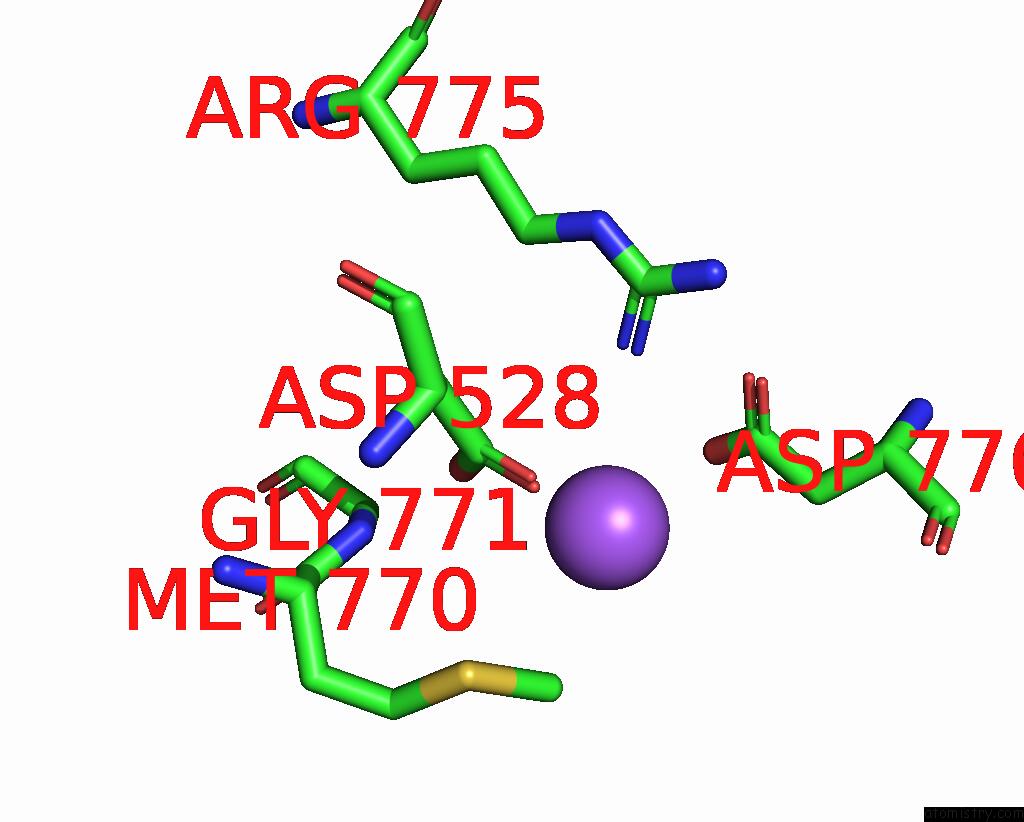
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 4 of Structure of the Ligand-Binding and Transmembrane Domains of Kainate Receptor GLUK2 in Complex with the Positive Allosteric Modulator BPAM344 and Noncompetitive Inhibitor Perampanel within 5.0Å range:
|
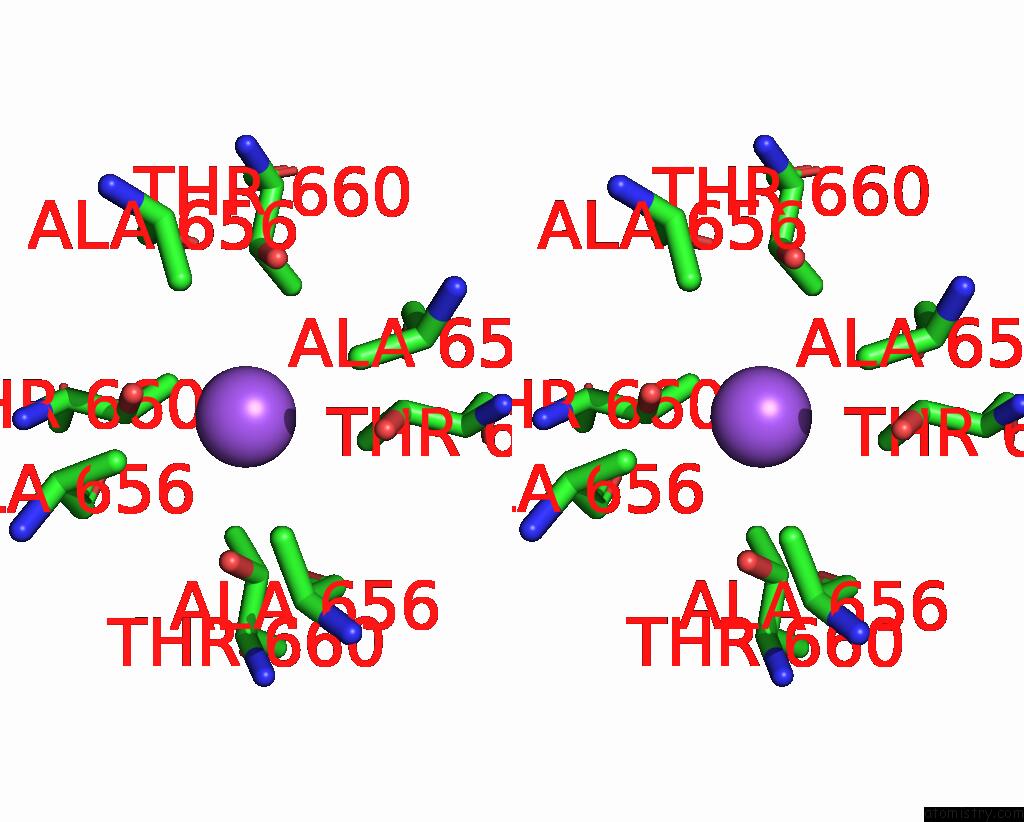
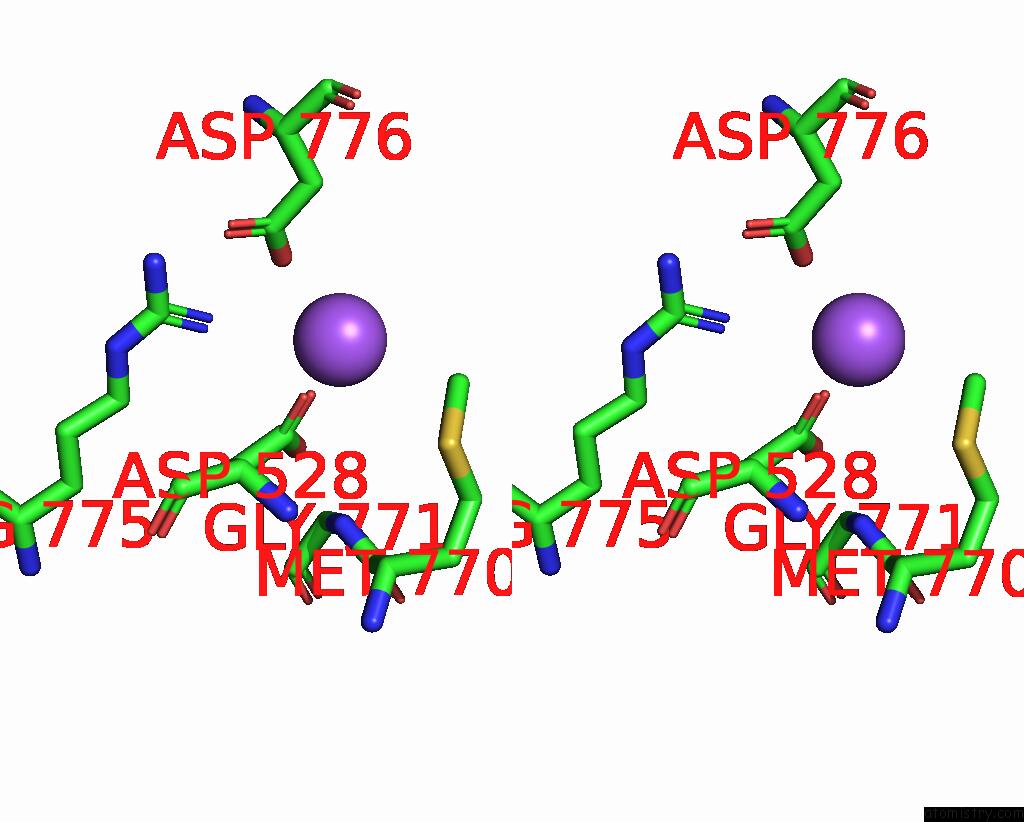
Sodium binding site 5 out of 7 in 8fww
Go back to
Sodium binding site 5 out
of 7 in the Structure of the Ligand-Binding and Transmembrane Domains of Kainate Receptor GLUK2 in Complex with the Positive Allosteric Modulator BPAM344 and Noncompetitive Inhibitor Perampanel

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 5 of Structure of the Ligand-Binding and Transmembrane Domains of Kainate Receptor GLUK2 in Complex with the Positive Allosteric Modulator BPAM344 and Noncompetitive Inhibitor Perampanel within 5.0Å range:
|
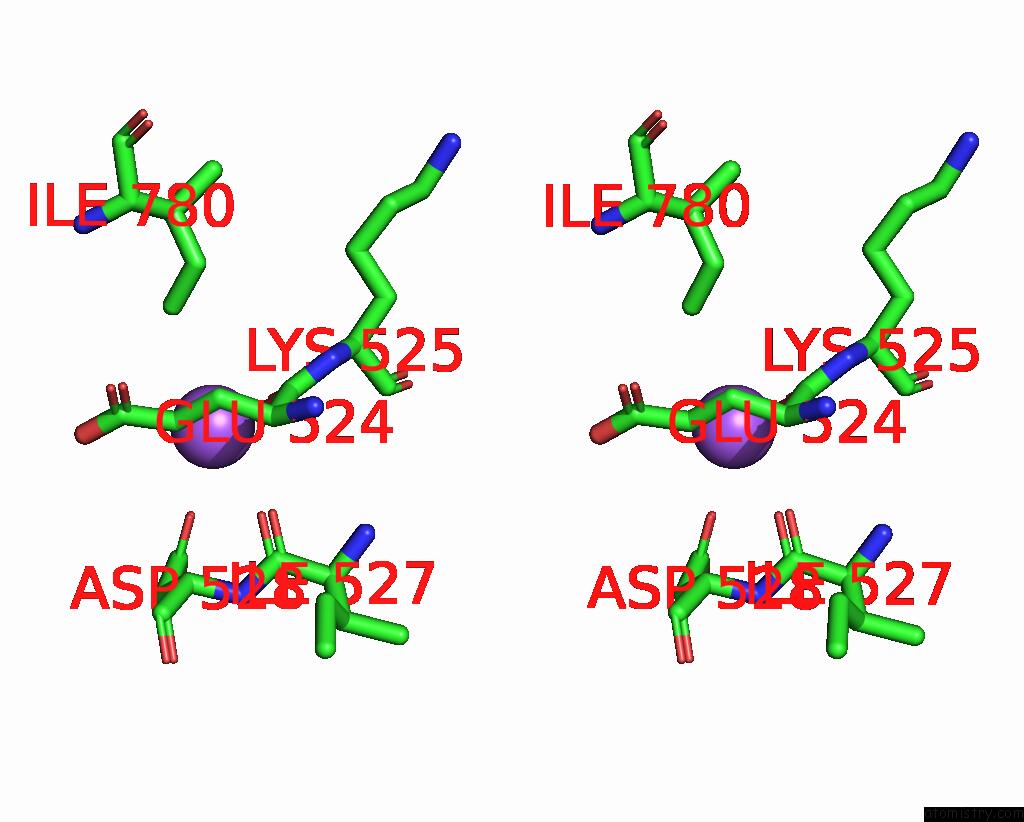
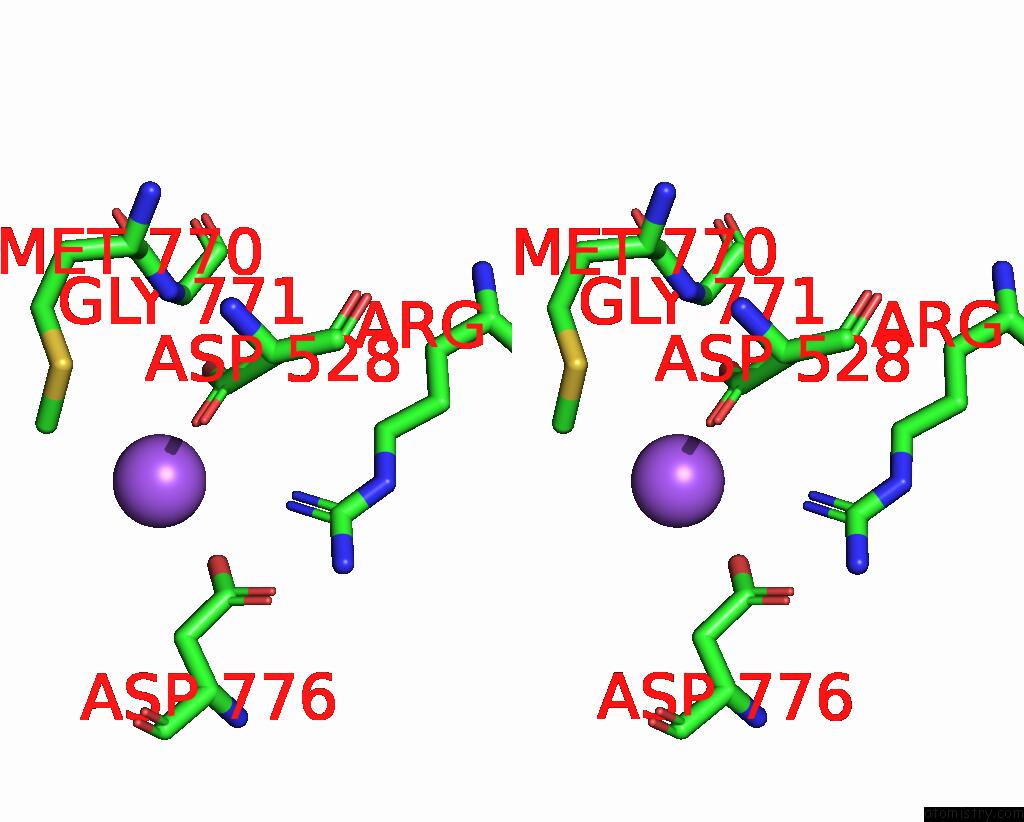
Sodium binding site 6 out of 7 in 8fww
Go back to
Sodium binding site 6 out
of 7 in the Structure of the Ligand-Binding and Transmembrane Domains of Kainate Receptor GLUK2 in Complex with the Positive Allosteric Modulator BPAM344 and Noncompetitive Inhibitor Perampanel
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 6 of Structure of the Ligand-Binding and Transmembrane Domains of Kainate Receptor GLUK2 in Complex with the Positive Allosteric Modulator BPAM344 and Noncompetitive Inhibitor Perampanel within 5.0Å range:
|
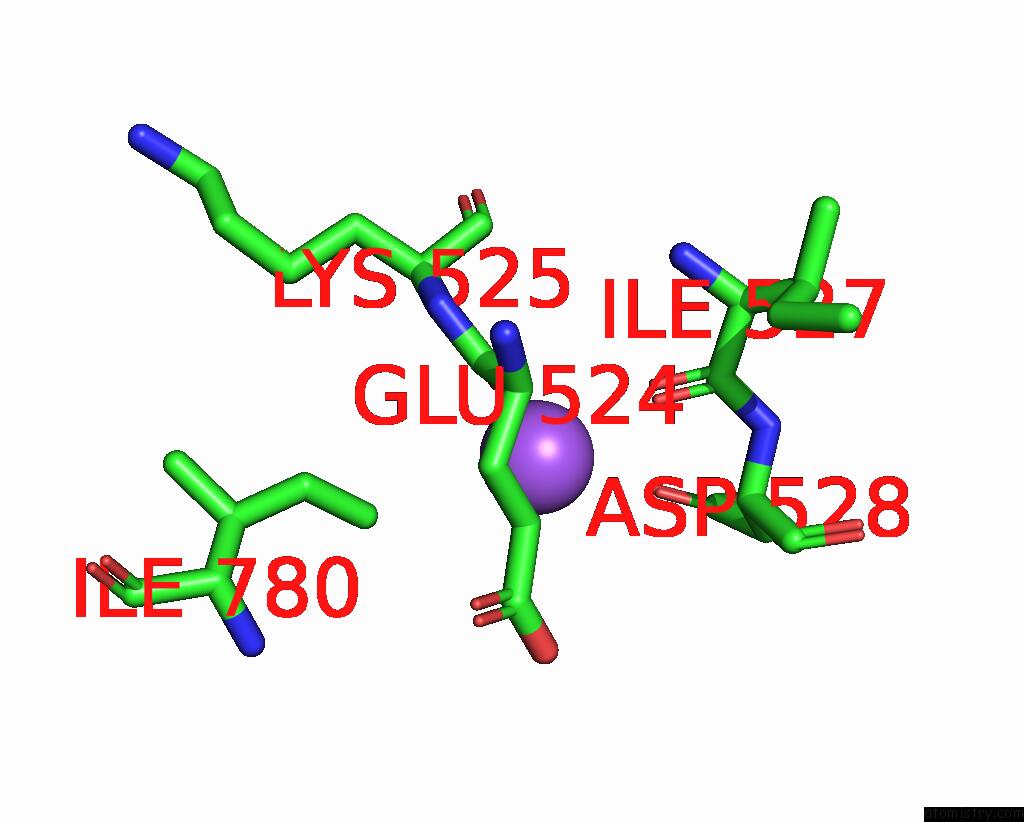
Sodium binding site 7 out of 7 in 8fww
Go back to
Sodium binding site 7 out
of 7 in the Structure of the Ligand-Binding and Transmembrane Domains of Kainate Receptor GLUK2 in Complex with the Positive Allosteric Modulator BPAM344 and Noncompetitive Inhibitor Perampanel
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 7 of Structure of the Ligand-Binding and Transmembrane Domains of Kainate Receptor GLUK2 in Complex with the Positive Allosteric Modulator BPAM344 and Noncompetitive Inhibitor Perampanel within 5.0Å range:
|
Reference:
S.P.Gangwar,
L.Y.Yen,
M.V.Yelshanskaya,
A.I.Sobolevsky.
Positive and Negative Allosteric Modulation of GLUK2 Kainate Receptors By BPAM344 and Antiepileptic Perampanel. Cell Rep V. 42 12124 2023.
ISSN: ESSN 2211-1247
PubMed: 36857176
DOI: 10.1016/J.CELREP.2023.112124
Page generated: Wed Oct 9 11:58:52 2024
ISSN: ESSN 2211-1247
PubMed: 36857176
DOI: 10.1016/J.CELREP.2023.112124
Last articles
Zn in 9MJ5Zn in 9HNW
Zn in 9G0L
Zn in 9FNE
Zn in 9DZN
Zn in 9E0I
Zn in 9D32
Zn in 9DAK
Zn in 8ZXC
Zn in 8ZUF