Sodium »
PDB 7a3i-7apu »
7a76 »
Sodium in PDB 7a76: Bacillithiol Disulfide Reductase Bdr (Ypda) From Bacillus Cereus
Enzymatic activity of Bacillithiol Disulfide Reductase Bdr (Ypda) From Bacillus Cereus
All present enzymatic activity of Bacillithiol Disulfide Reductase Bdr (Ypda) From Bacillus Cereus:
1.8.1.9;
1.8.1.9;
Protein crystallography data
The structure of Bacillithiol Disulfide Reductase Bdr (Ypda) From Bacillus Cereus, PDB code: 7a76
was solved by
M.Hammerstad,
I.Gudim,
H.-P.Hersleth,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.23 / 1.65 |
| Space group | P 2 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 88.731, 114.9, 146.73, 90, 90, 90 |
| R / Rfree (%) | 19 / 21.8 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Bacillithiol Disulfide Reductase Bdr (Ypda) From Bacillus Cereus
(pdb code 7a76). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 4 binding sites of Sodium where determined in the Bacillithiol Disulfide Reductase Bdr (Ypda) From Bacillus Cereus, PDB code: 7a76:
Jump to Sodium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Sodium where determined in the Bacillithiol Disulfide Reductase Bdr (Ypda) From Bacillus Cereus, PDB code: 7a76:
Jump to Sodium binding site number: 1; 2; 3; 4;
Sodium binding site 1 out of 4 in 7a76
Go back to
Sodium binding site 1 out
of 4 in the Bacillithiol Disulfide Reductase Bdr (Ypda) From Bacillus Cereus
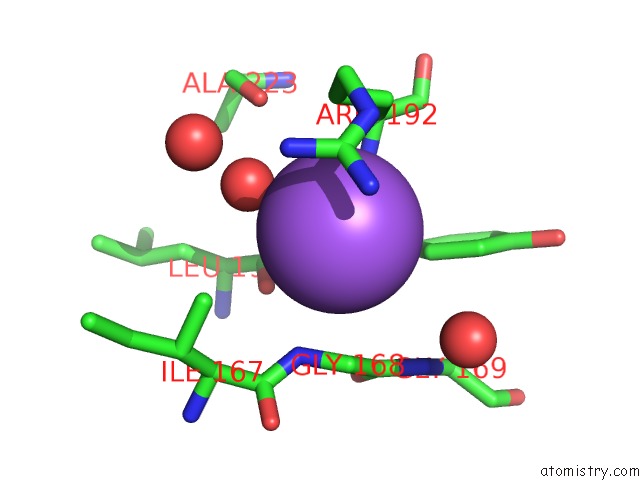
Mono view
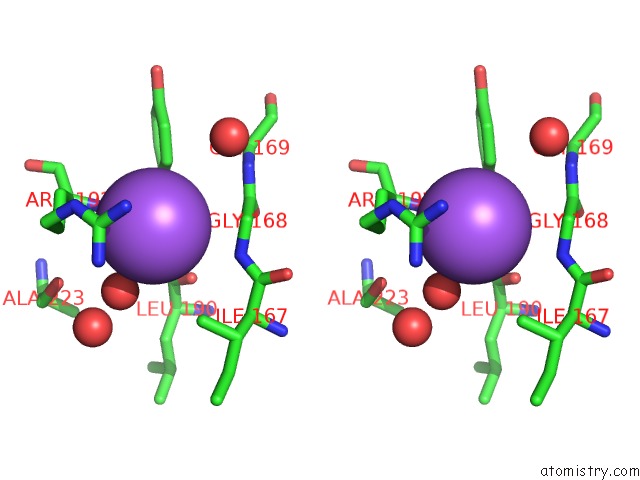
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Bacillithiol Disulfide Reductase Bdr (Ypda) From Bacillus Cereus within 5.0Å range:
|
Sodium binding site 2 out of 4 in 7a76
Go back to
Sodium binding site 2 out
of 4 in the Bacillithiol Disulfide Reductase Bdr (Ypda) From Bacillus Cereus
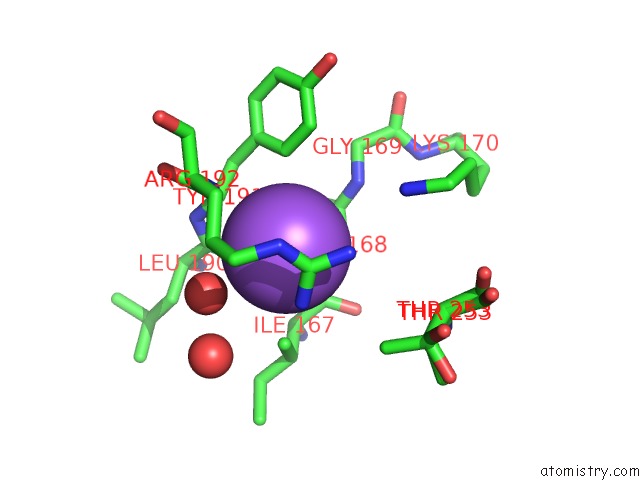
Mono view
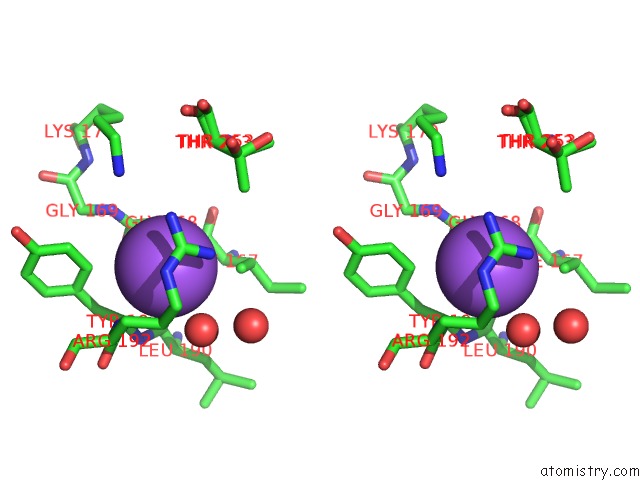
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Bacillithiol Disulfide Reductase Bdr (Ypda) From Bacillus Cereus within 5.0Å range:
|
Sodium binding site 3 out of 4 in 7a76
Go back to
Sodium binding site 3 out
of 4 in the Bacillithiol Disulfide Reductase Bdr (Ypda) From Bacillus Cereus
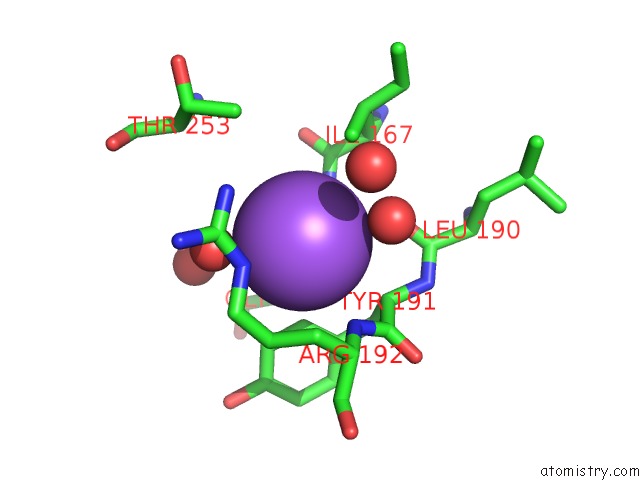
Mono view
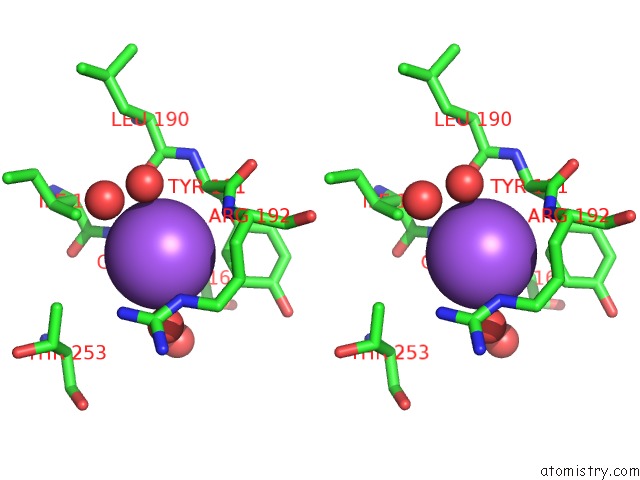
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 3 of Bacillithiol Disulfide Reductase Bdr (Ypda) From Bacillus Cereus within 5.0Å range:
|
Sodium binding site 4 out of 4 in 7a76
Go back to
Sodium binding site 4 out
of 4 in the Bacillithiol Disulfide Reductase Bdr (Ypda) From Bacillus Cereus
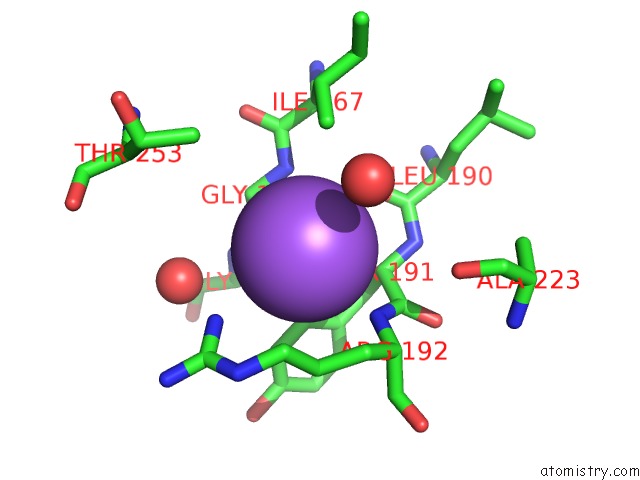
Mono view
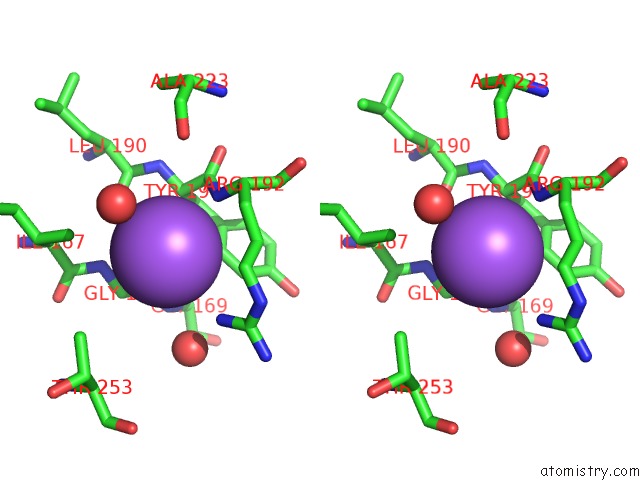
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 4 of Bacillithiol Disulfide Reductase Bdr (Ypda) From Bacillus Cereus within 5.0Å range:
|
Reference:
M.Hammerstad,
I.Gudim,
H.P.Hersleth.
The Crystal Structures of Bacillithiol Disulfide Reductase Bdr (Ypda) Provide Structural and Functional Insight Into A New Type of Fad-Containing Nadph-Dependent Oxidoreductase. Biochemistry V. 59 4793 2020.
ISSN: ISSN 0006-2960
PubMed: 33326741
DOI: 10.1021/ACS.BIOCHEM.0C00745
Page generated: Mon Aug 18 09:02:24 2025
ISSN: ISSN 0006-2960
PubMed: 33326741
DOI: 10.1021/ACS.BIOCHEM.0C00745
Last articles
Na in 7SLXNa in 7SKG
Na in 7SLV
Na in 7SKF
Na in 7SKE
Na in 7SKD
Na in 7SJW
Na in 7SJV
Na in 7SJU
Na in 7SJT