Sodium »
PDB 6syr-6tcj »
6tb1 »
Sodium in PDB 6tb1: Crystal Structure of Thermostable Omega Transaminase 6-Fold Mutant From Pseudomonas Jessenii
Protein crystallography data
The structure of Crystal Structure of Thermostable Omega Transaminase 6-Fold Mutant From Pseudomonas Jessenii, PDB code: 6tb1
was solved by
N.Capra,
H.J.Rozeboom,
A.M.W.H.Thunnissen,
D.B.Janssen,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.74 / 1.85 |
| Space group | P 43 |
| Cell size a, b, c (Å), α, β, γ (°) | 98.924, 98.924, 120.276, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 15.6 / 18.7 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Crystal Structure of Thermostable Omega Transaminase 6-Fold Mutant From Pseudomonas Jessenii
(pdb code 6tb1). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 2 binding sites of Sodium where determined in the Crystal Structure of Thermostable Omega Transaminase 6-Fold Mutant From Pseudomonas Jessenii, PDB code: 6tb1:
Jump to Sodium binding site number: 1; 2;
In total 2 binding sites of Sodium where determined in the Crystal Structure of Thermostable Omega Transaminase 6-Fold Mutant From Pseudomonas Jessenii, PDB code: 6tb1:
Jump to Sodium binding site number: 1; 2;
Sodium binding site 1 out of 2 in 6tb1
Go back to
Sodium binding site 1 out
of 2 in the Crystal Structure of Thermostable Omega Transaminase 6-Fold Mutant From Pseudomonas Jessenii
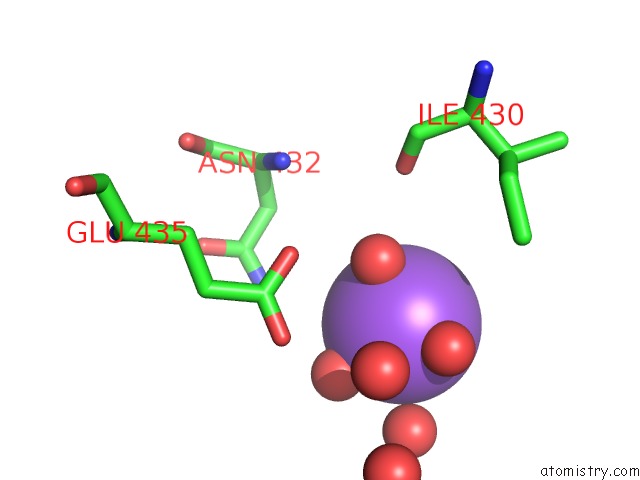
Mono view
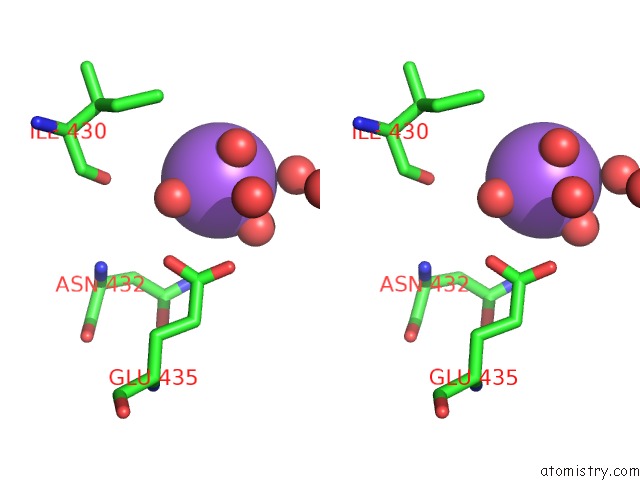
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Crystal Structure of Thermostable Omega Transaminase 6-Fold Mutant From Pseudomonas Jessenii within 5.0Å range:
|
Sodium binding site 2 out of 2 in 6tb1
Go back to
Sodium binding site 2 out
of 2 in the Crystal Structure of Thermostable Omega Transaminase 6-Fold Mutant From Pseudomonas Jessenii
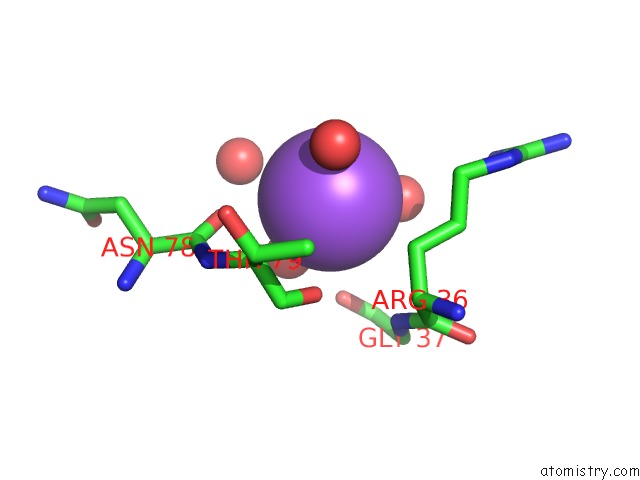
Mono view
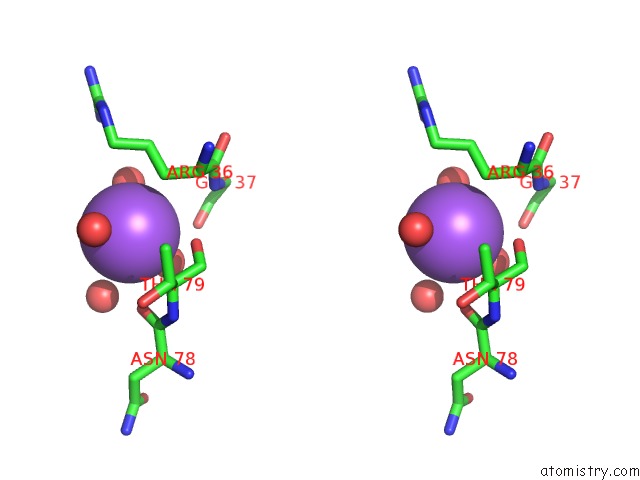
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Crystal Structure of Thermostable Omega Transaminase 6-Fold Mutant From Pseudomonas Jessenii within 5.0Å range:
|
Reference:
Q.Meng,
N.Capra,
C.M.Palacio,
E.Lanfranchi,
M.Otzen,
L.Z.Van Schie,
H.J.Rozeboom,
A.W.H.Thunnissen,
H.J.Wijma,
D.B.Janssen.
Robust Omega-Transaminases By Computational Stabilization of the Subunit Interface. Acs Catalysis V. 10 2915 2020.
ISSN: ESSN 2155-5435
PubMed: 32953233
DOI: 10.1021/ACSCATAL.9B05223
Page generated: Mon Aug 18 07:19:49 2025
ISSN: ESSN 2155-5435
PubMed: 32953233
DOI: 10.1021/ACSCATAL.9B05223
Last articles
Zn in 3KM2Zn in 3KL9
Zn in 3KQI
Zn in 3KQ6
Zn in 3KNX
Zn in 3KNV
Zn in 3KNS
Zn in 3KNR
Zn in 3KNE
Zn in 3KN2