Sodium »
PDB 6syr-6tcj »
6t46 »
Sodium in PDB 6t46: Structure of the Rap Conjugation Gene Regulator of the Plasmid PLS20 in Complex with the Phr* Peptide
Protein crystallography data
The structure of Structure of the Rap Conjugation Gene Regulator of the Plasmid PLS20 in Complex with the Phr* Peptide, PDB code: 6t46
was solved by
I.Crespo,
N.Bernardo,
W.J.J.Meijer,
D.R.Boer,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.42 / 2.45 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 116.670, 93.282, 167.687, 90.00, 94.97, 90.00 |
| R / Rfree (%) | 20.2 / 26.2 |
Other elements in 6t46:
The structure of Structure of the Rap Conjugation Gene Regulator of the Plasmid PLS20 in Complex with the Phr* Peptide also contains other interesting chemical elements:
| Chlorine | (Cl) | 3 atoms |
Sodium Binding Sites:
The binding sites of Sodium atom in the Structure of the Rap Conjugation Gene Regulator of the Plasmid PLS20 in Complex with the Phr* Peptide
(pdb code 6t46). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Structure of the Rap Conjugation Gene Regulator of the Plasmid PLS20 in Complex with the Phr* Peptide, PDB code: 6t46:
In total only one binding site of Sodium was determined in the Structure of the Rap Conjugation Gene Regulator of the Plasmid PLS20 in Complex with the Phr* Peptide, PDB code: 6t46:
Sodium binding site 1 out of 1 in 6t46
Go back to
Sodium binding site 1 out
of 1 in the Structure of the Rap Conjugation Gene Regulator of the Plasmid PLS20 in Complex with the Phr* Peptide
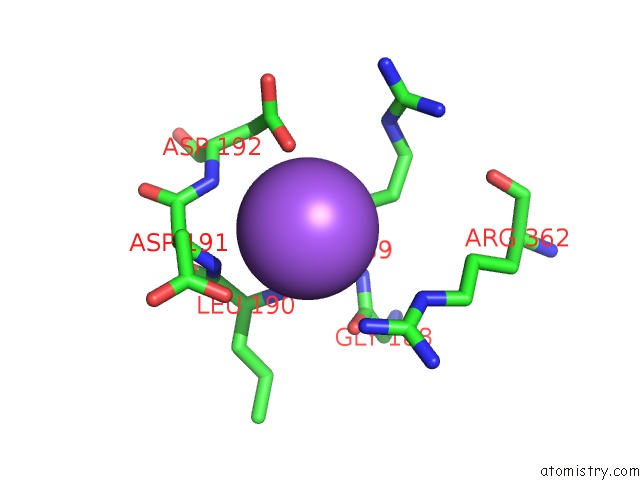
Mono view
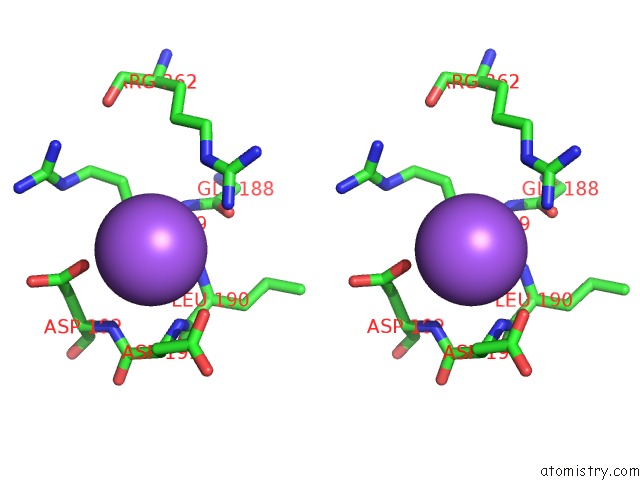
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Structure of the Rap Conjugation Gene Regulator of the Plasmid PLS20 in Complex with the Phr* Peptide within 5.0Å range:
|
Reference:
I.Crespo,
N.Bernardo,
A.Miguel-Arribas,
P.K.Singh,
J.R.Luque-Ortega,
C.Alfonso,
M.Malfois,
W.J.J.Meijer,
D.R.Boer.
Inactivation of the Dimeric RAPPLS20 Anti-Repressor of the Conjugation Operon Is Mediated By Signaling Peptide-Induced Tetramerization Nucleic Acids Res. 2020.
ISSN: ESSN 1362-4962
Page generated: Mon Aug 18 07:17:04 2025
ISSN: ESSN 1362-4962
Last articles
Zn in 3JC6Zn in 3JBX
Zn in 3JAP
Zn in 3JBW
Zn in 3JAM
Zn in 3J0K
Zn in 3J9S
Zn in 3J81
Zn in 3J9D
Zn in 3J80