Sodium »
PDB 5oqw-5pol »
5ou5 »
Sodium in PDB 5ou5: Crystal Structure of Maize Chloroplastic Photosynthetic Nadp(+)- Dependent Malic Enzyme
Protein crystallography data
The structure of Crystal Structure of Maize Chloroplastic Photosynthetic Nadp(+)- Dependent Malic Enzyme, PDB code: 5ou5
was solved by
A.Bovdilova,
A.Hoeppner,
V.G.Maurino,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.88 / 2.20 |
| Space group | I 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 135.961, 147.155, 261.615, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16 / 21.1 |
Other elements in 5ou5:
The structure of Crystal Structure of Maize Chloroplastic Photosynthetic Nadp(+)- Dependent Malic Enzyme also contains other interesting chemical elements:
| Potassium | (K) | 1 atom |
Sodium Binding Sites:
Pages:
>>> Page 1 <<< Page 2, Binding sites: 11 - 16;Binding sites:
The binding sites of Sodium atom in the Crystal Structure of Maize Chloroplastic Photosynthetic Nadp(+)- Dependent Malic Enzyme (pdb code 5ou5). This binding sites where shown within 5.0 Angstroms radius around Sodium atom.In total 16 binding sites of Sodium where determined in the Crystal Structure of Maize Chloroplastic Photosynthetic Nadp(+)- Dependent Malic Enzyme, PDB code: 5ou5:
Jump to Sodium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Sodium binding site 1 out of 16 in 5ou5
Go back to
Sodium binding site 1 out
of 16 in the Crystal Structure of Maize Chloroplastic Photosynthetic Nadp(+)- Dependent Malic Enzyme
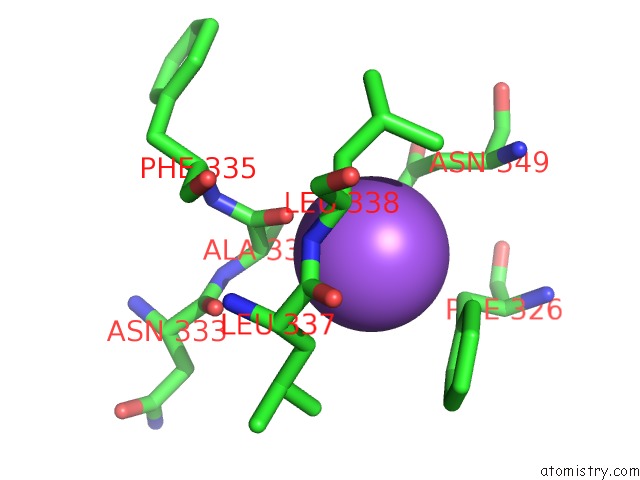
Mono view
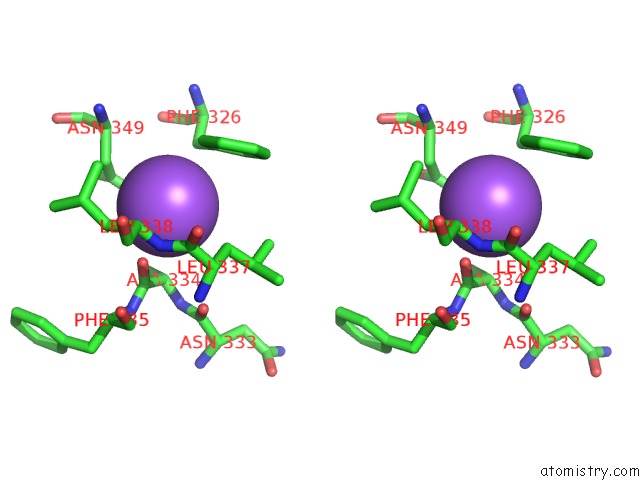
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Crystal Structure of Maize Chloroplastic Photosynthetic Nadp(+)- Dependent Malic Enzyme within 5.0Å range:
|
Sodium binding site 2 out of 16 in 5ou5
Go back to
Sodium binding site 2 out
of 16 in the Crystal Structure of Maize Chloroplastic Photosynthetic Nadp(+)- Dependent Malic Enzyme
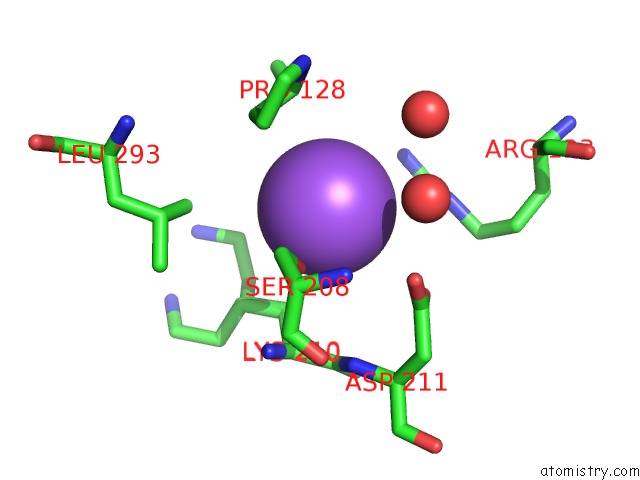
Mono view
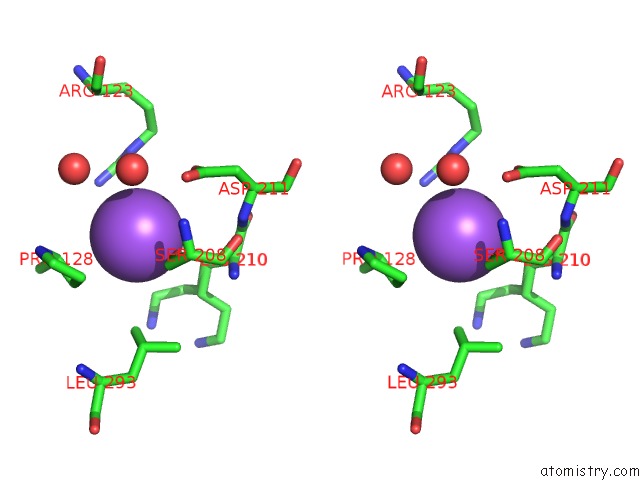
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Crystal Structure of Maize Chloroplastic Photosynthetic Nadp(+)- Dependent Malic Enzyme within 5.0Å range:
|
Sodium binding site 3 out of 16 in 5ou5
Go back to
Sodium binding site 3 out
of 16 in the Crystal Structure of Maize Chloroplastic Photosynthetic Nadp(+)- Dependent Malic Enzyme
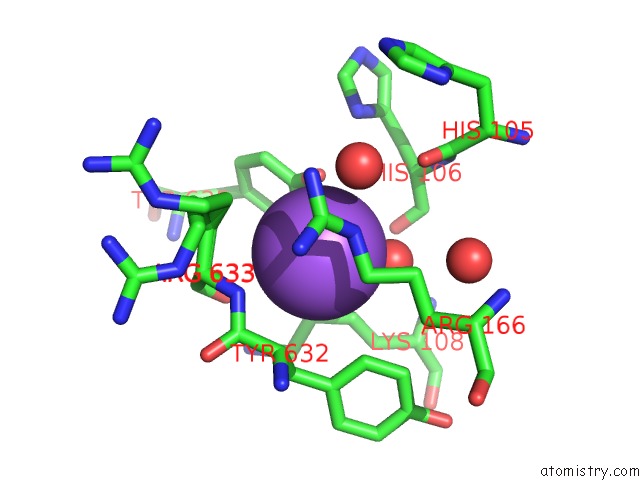
Mono view
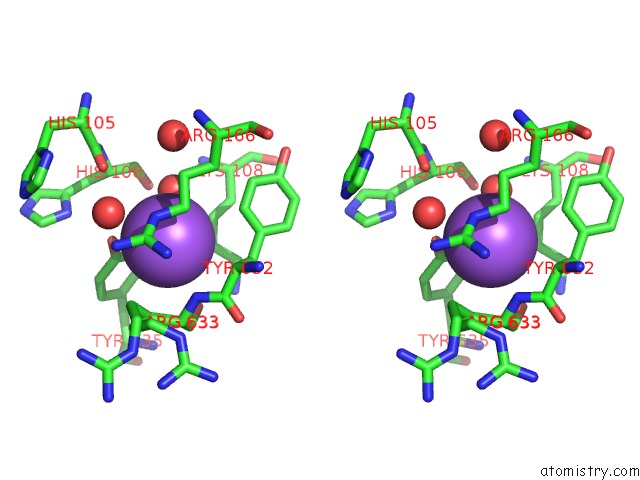
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 3 of Crystal Structure of Maize Chloroplastic Photosynthetic Nadp(+)- Dependent Malic Enzyme within 5.0Å range:
|
Sodium binding site 4 out of 16 in 5ou5
Go back to
Sodium binding site 4 out
of 16 in the Crystal Structure of Maize Chloroplastic Photosynthetic Nadp(+)- Dependent Malic Enzyme
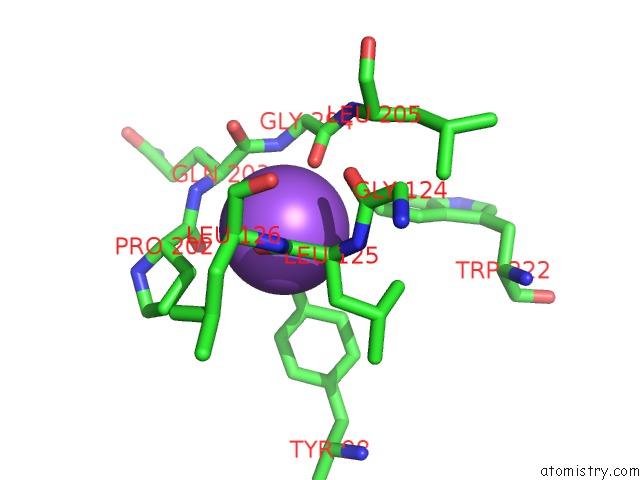
Mono view
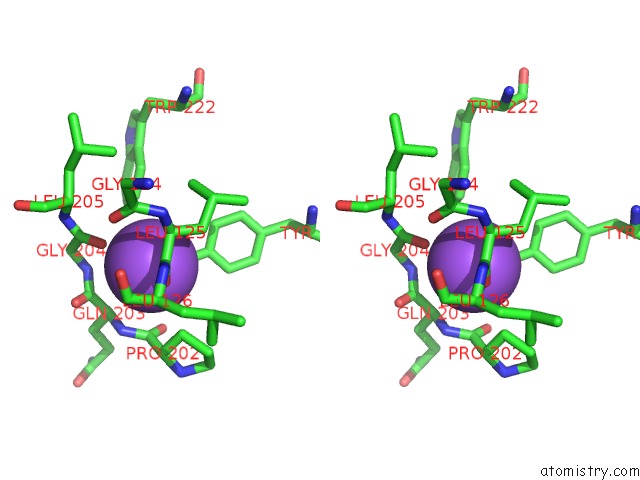
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 4 of Crystal Structure of Maize Chloroplastic Photosynthetic Nadp(+)- Dependent Malic Enzyme within 5.0Å range:
|
Sodium binding site 5 out of 16 in 5ou5
Go back to
Sodium binding site 5 out
of 16 in the Crystal Structure of Maize Chloroplastic Photosynthetic Nadp(+)- Dependent Malic Enzyme
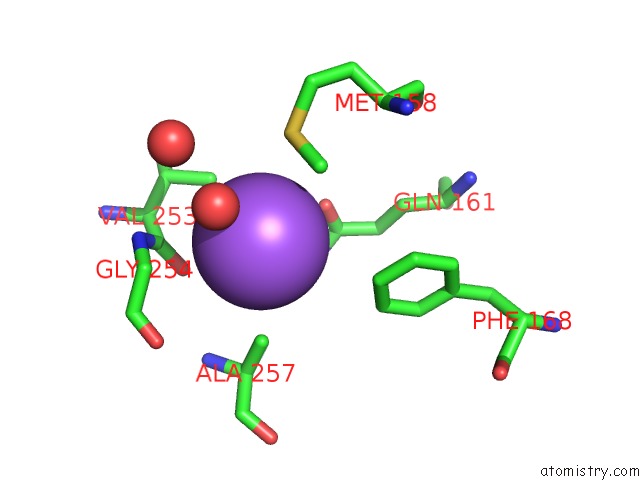
Mono view
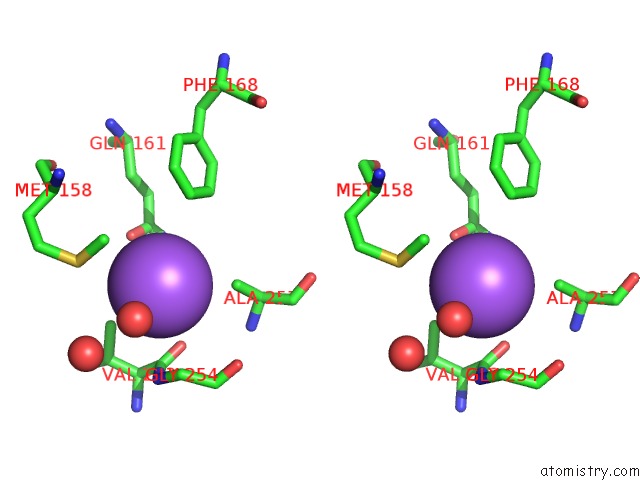
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 5 of Crystal Structure of Maize Chloroplastic Photosynthetic Nadp(+)- Dependent Malic Enzyme within 5.0Å range:
|
Sodium binding site 6 out of 16 in 5ou5
Go back to
Sodium binding site 6 out
of 16 in the Crystal Structure of Maize Chloroplastic Photosynthetic Nadp(+)- Dependent Malic Enzyme
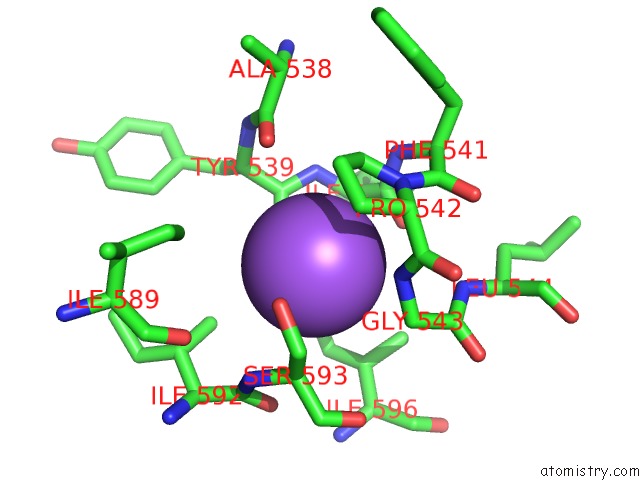
Mono view
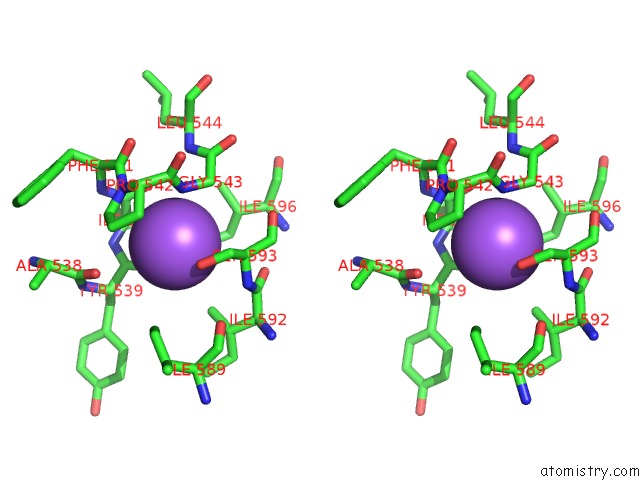
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 6 of Crystal Structure of Maize Chloroplastic Photosynthetic Nadp(+)- Dependent Malic Enzyme within 5.0Å range:
|
Sodium binding site 7 out of 16 in 5ou5
Go back to
Sodium binding site 7 out
of 16 in the Crystal Structure of Maize Chloroplastic Photosynthetic Nadp(+)- Dependent Malic Enzyme
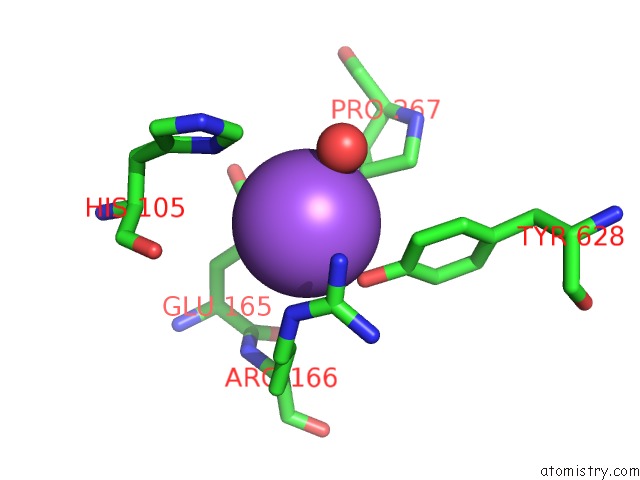
Mono view
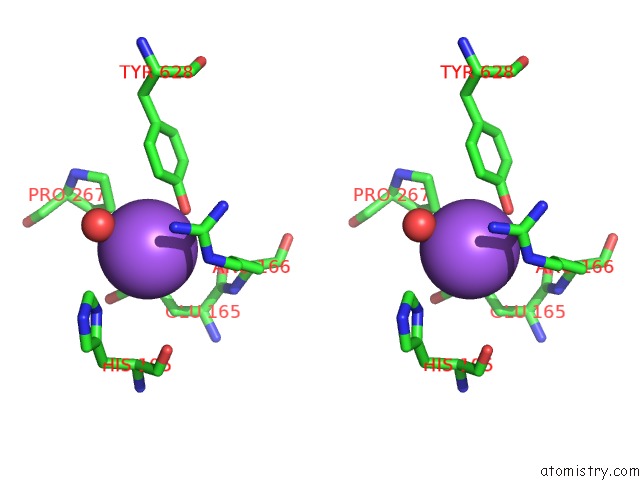
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 7 of Crystal Structure of Maize Chloroplastic Photosynthetic Nadp(+)- Dependent Malic Enzyme within 5.0Å range:
|
Sodium binding site 8 out of 16 in 5ou5
Go back to
Sodium binding site 8 out
of 16 in the Crystal Structure of Maize Chloroplastic Photosynthetic Nadp(+)- Dependent Malic Enzyme
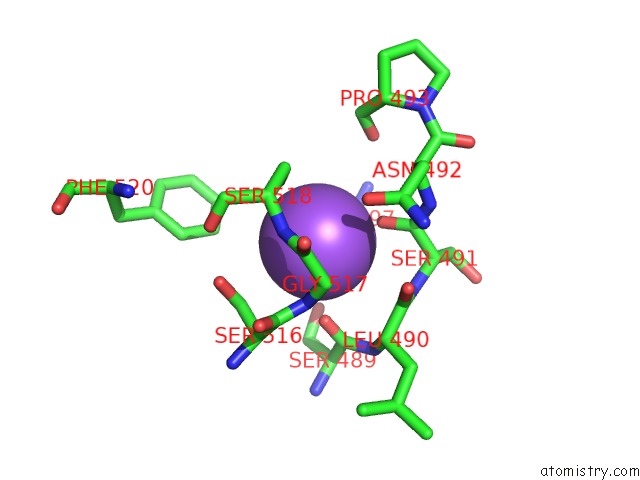
Mono view
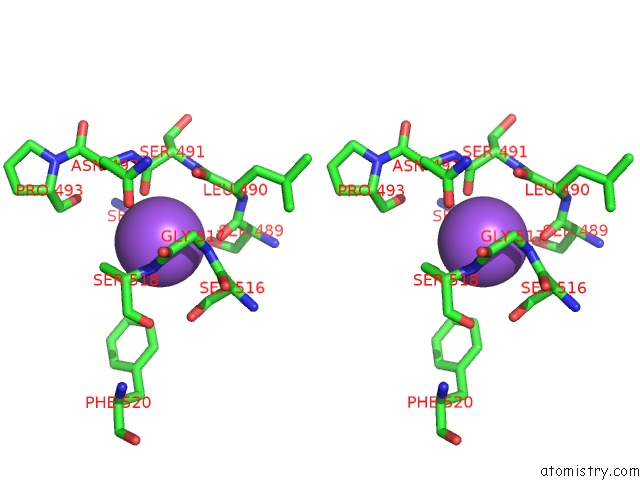
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 8 of Crystal Structure of Maize Chloroplastic Photosynthetic Nadp(+)- Dependent Malic Enzyme within 5.0Å range:
|
Sodium binding site 9 out of 16 in 5ou5
Go back to
Sodium binding site 9 out
of 16 in the Crystal Structure of Maize Chloroplastic Photosynthetic Nadp(+)- Dependent Malic Enzyme
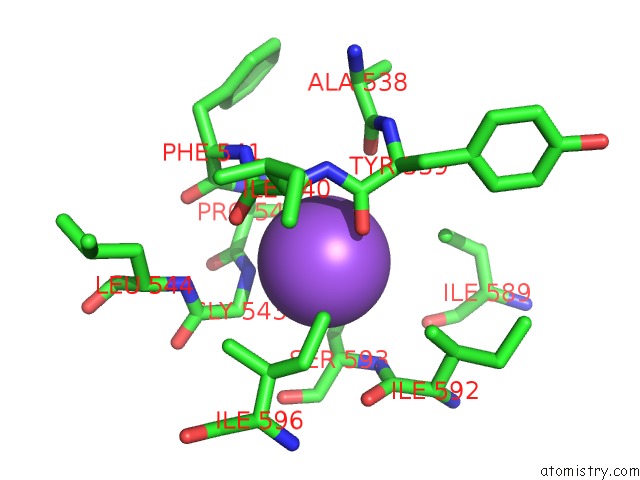
Mono view
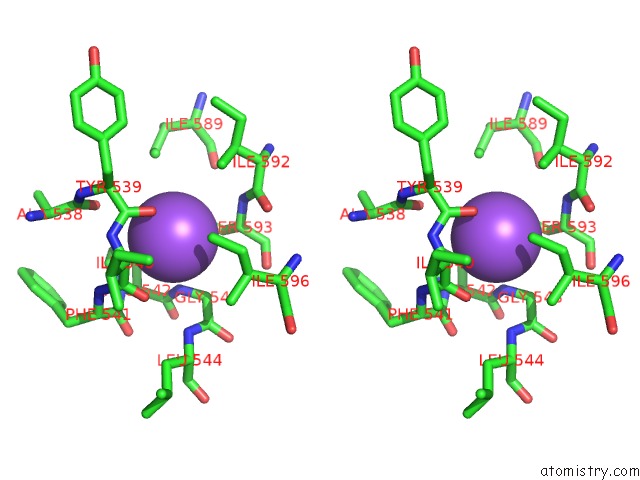
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 9 of Crystal Structure of Maize Chloroplastic Photosynthetic Nadp(+)- Dependent Malic Enzyme within 5.0Å range:
|
Sodium binding site 10 out of 16 in 5ou5
Go back to
Sodium binding site 10 out
of 16 in the Crystal Structure of Maize Chloroplastic Photosynthetic Nadp(+)- Dependent Malic Enzyme
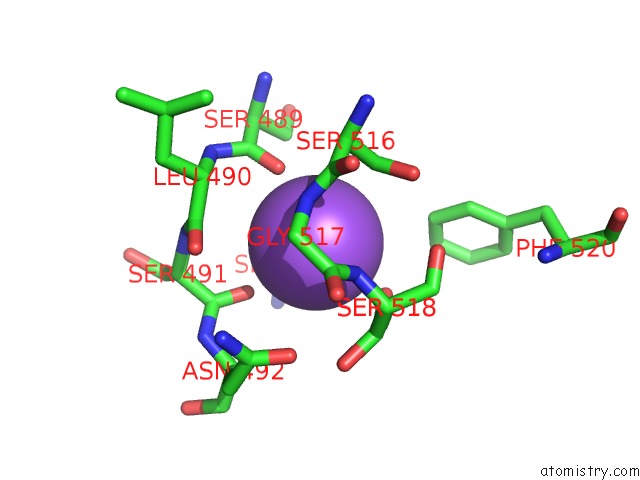
Mono view
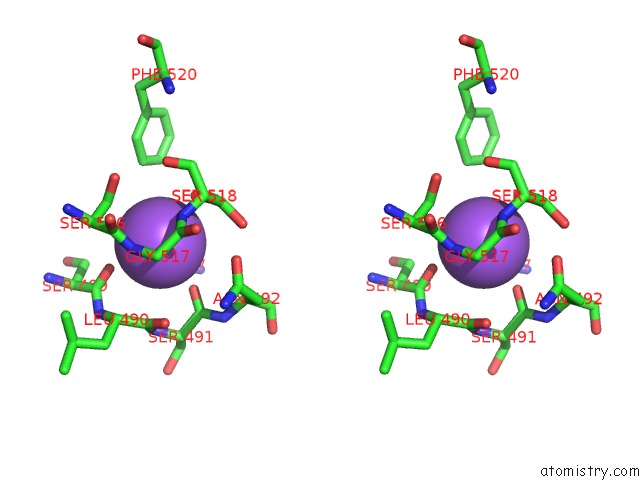
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 10 of Crystal Structure of Maize Chloroplastic Photosynthetic Nadp(+)- Dependent Malic Enzyme within 5.0Å range:
|
Reference:
C.E.Alvarez,
A.Bovdilova,
A.Hoppner,
C.C.Wolff,
M.Saigo,
F.Trajtenberg,
T.Zhang,
A.Buschiazzo,
L.Nagel-Steger,
M.F.Drincovich,
M.J.Lercher,
V.G.Maurino.
Molecular Adaptations of Nadp-Malic Enzyme For Its Function in C4PHOTOSYNTHESIS in Grasses. Nat.Plants V. 5 755 2019.
ISSN: ESSN 2055-0278
PubMed: 31235877
DOI: 10.1038/S41477-019-0451-7
Page generated: Mon Oct 7 23:15:07 2024
ISSN: ESSN 2055-0278
PubMed: 31235877
DOI: 10.1038/S41477-019-0451-7
Last articles
Cl in 5GZXCl in 5H22
Cl in 5GY0
Cl in 5H0Z
Cl in 5H0Y
Cl in 5GZK
Cl in 5GXZ
Cl in 5GWF
Cl in 5GXY
Cl in 5GXX