Sodium »
PDB 5mss-5nbj »
5n84 »
Sodium in PDB 5n84: Ttk Kinase Domain in Complex with Mps-BAY2B
Enzymatic activity of Ttk Kinase Domain in Complex with Mps-BAY2B
All present enzymatic activity of Ttk Kinase Domain in Complex with Mps-BAY2B:
2.7.12.1;
2.7.12.1;
Protein crystallography data
The structure of Ttk Kinase Domain in Complex with Mps-BAY2B, PDB code: 5n84
was solved by
J.Uitdehaag,
N.Willemsen-Seegers,
J.De Man,
R.C.Buijsman,
G.J.R.Zaman,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 41.73 / 2.30 |
| Space group | I 2 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 71.178, 112.199, 115.983, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.5 / 26.1 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Ttk Kinase Domain in Complex with Mps-BAY2B
(pdb code 5n84). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 5 binding sites of Sodium where determined in the Ttk Kinase Domain in Complex with Mps-BAY2B, PDB code: 5n84:
Jump to Sodium binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Sodium where determined in the Ttk Kinase Domain in Complex with Mps-BAY2B, PDB code: 5n84:
Jump to Sodium binding site number: 1; 2; 3; 4; 5;
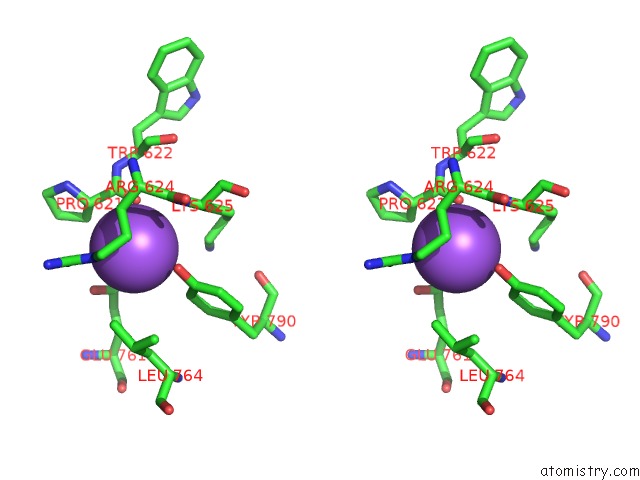
Sodium binding site 1 out of 5 in 5n84
Go back to
Sodium binding site 1 out
of 5 in the Ttk Kinase Domain in Complex with Mps-BAY2B

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Ttk Kinase Domain in Complex with Mps-BAY2B within 5.0Å range:
|
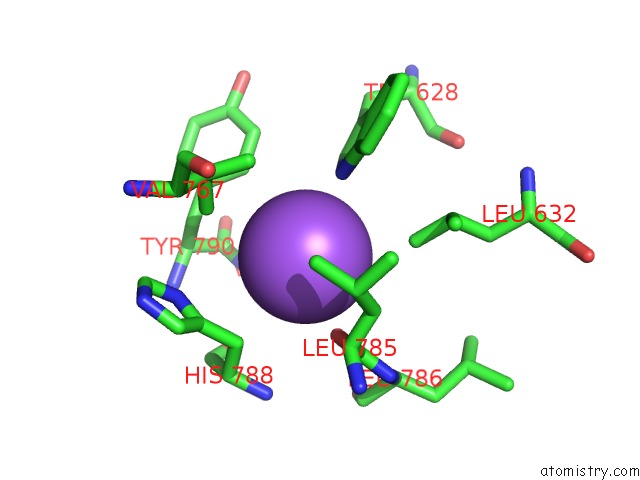
Sodium binding site 2 out of 5 in 5n84
Go back to
Sodium binding site 2 out
of 5 in the Ttk Kinase Domain in Complex with Mps-BAY2B
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Ttk Kinase Domain in Complex with Mps-BAY2B within 5.0Å range:
|
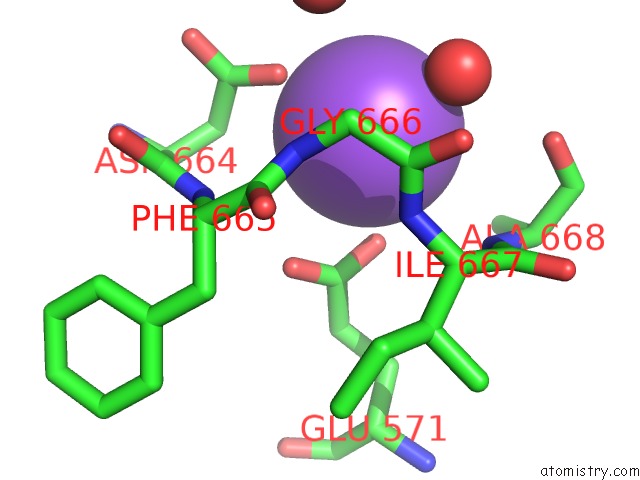
Sodium binding site 3 out of 5 in 5n84
Go back to
Sodium binding site 3 out
of 5 in the Ttk Kinase Domain in Complex with Mps-BAY2B
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 3 of Ttk Kinase Domain in Complex with Mps-BAY2B within 5.0Å range:
|
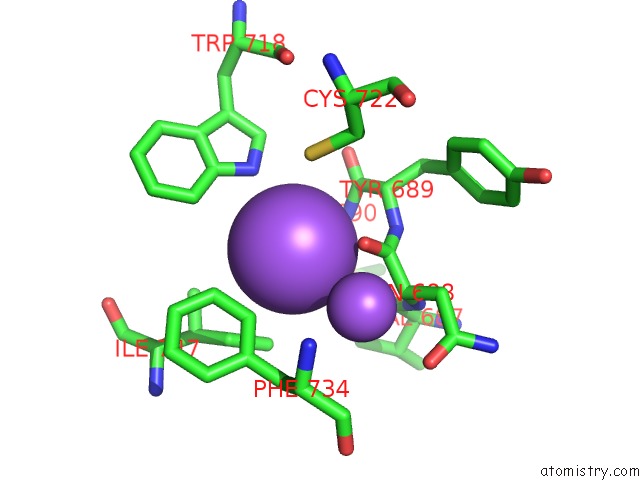
Sodium binding site 4 out of 5 in 5n84
Go back to
Sodium binding site 4 out
of 5 in the Ttk Kinase Domain in Complex with Mps-BAY2B
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 4 of Ttk Kinase Domain in Complex with Mps-BAY2B within 5.0Å range:
|
Sodium binding site 5 out of 5 in 5n84
Go back to
Sodium binding site 5 out
of 5 in the Ttk Kinase Domain in Complex with Mps-BAY2B

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 5 of Ttk Kinase Domain in Complex with Mps-BAY2B within 5.0Å range:
|
Reference:
J.C.M.Uitdehaag,
J.De Man,
N.Willemsen-Seegers,
M.B.W.Prinsen,
M.A.A.Libouban,
J.G.Sterrenburg,
J.J.P.De Wit,
J.R.F.De Vetter,
J.A.D.M.De Roos,
R.C.Buijsman,
G.J.R.Zaman.
Target Residence Time-Guided Optimization on Ttk Kinase Results in Inhibitors with Potent Anti-Proliferative Activity. J. Mol. Biol. V. 429 2211 2017.
ISSN: ESSN 1089-8638
PubMed: 28539250
DOI: 10.1016/J.JMB.2017.05.014
Page generated: Mon Oct 7 22:51:17 2024
ISSN: ESSN 1089-8638
PubMed: 28539250
DOI: 10.1016/J.JMB.2017.05.014
Last articles
Cl in 5T8WCl in 5T8Q
Cl in 5T77
Cl in 5T8I
Cl in 5T8E
Cl in 5T7U
Cl in 5T7F
Cl in 5T7P
Cl in 5T7T
Cl in 5T79