Sodium »
PDB 5m1y-5mfs »
5m6i »
Sodium in PDB 5m6i: Crystal Structure of Non-Cardiotoxic Bence-Jones Light Chain Dimer M8
Protein crystallography data
The structure of Crystal Structure of Non-Cardiotoxic Bence-Jones Light Chain Dimer M8, PDB code: 5m6i
was solved by
L.Oberti,
P.Rognoni,
R.Russo,
J.Bacarizo,
M.Bolognesi,
S.Ricagno,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 54.56 / 2.20 |
| Space group | I 4 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 229.378, 229.378, 64.429, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19.5 / 22.7 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Crystal Structure of Non-Cardiotoxic Bence-Jones Light Chain Dimer M8
(pdb code 5m6i). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 2 binding sites of Sodium where determined in the Crystal Structure of Non-Cardiotoxic Bence-Jones Light Chain Dimer M8, PDB code: 5m6i:
Jump to Sodium binding site number: 1; 2;
In total 2 binding sites of Sodium where determined in the Crystal Structure of Non-Cardiotoxic Bence-Jones Light Chain Dimer M8, PDB code: 5m6i:
Jump to Sodium binding site number: 1; 2;
Sodium binding site 1 out of 2 in 5m6i
Go back to
Sodium binding site 1 out
of 2 in the Crystal Structure of Non-Cardiotoxic Bence-Jones Light Chain Dimer M8
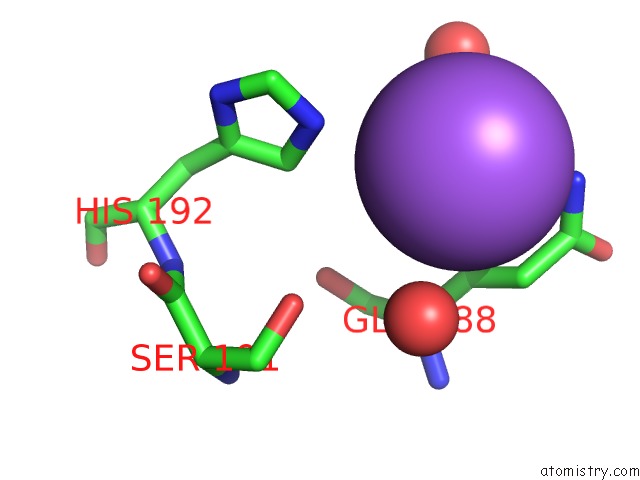
Mono view
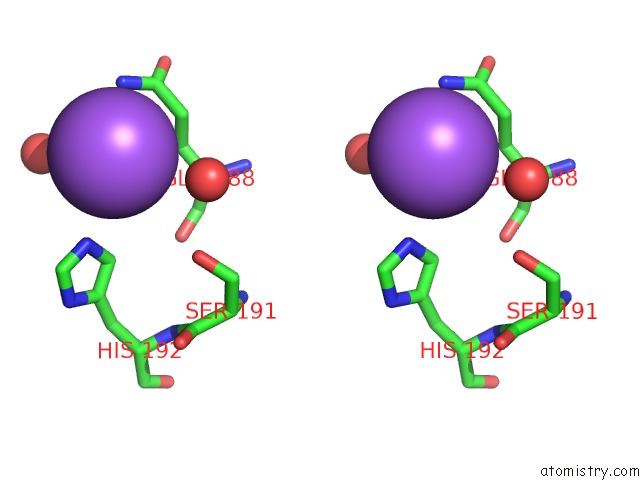
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Crystal Structure of Non-Cardiotoxic Bence-Jones Light Chain Dimer M8 within 5.0Å range:
|
Sodium binding site 2 out of 2 in 5m6i
Go back to
Sodium binding site 2 out
of 2 in the Crystal Structure of Non-Cardiotoxic Bence-Jones Light Chain Dimer M8
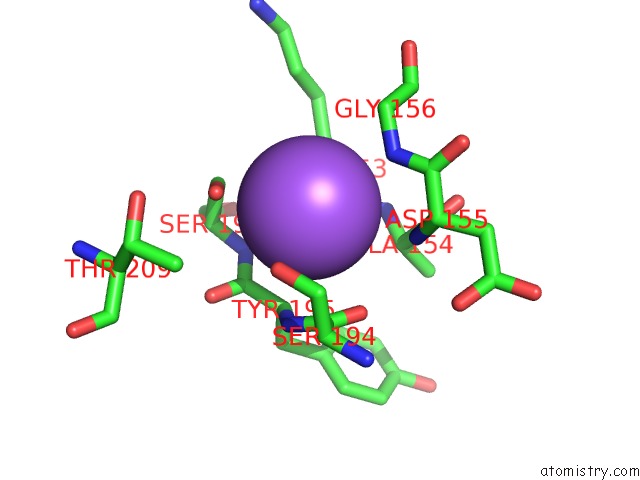
Mono view
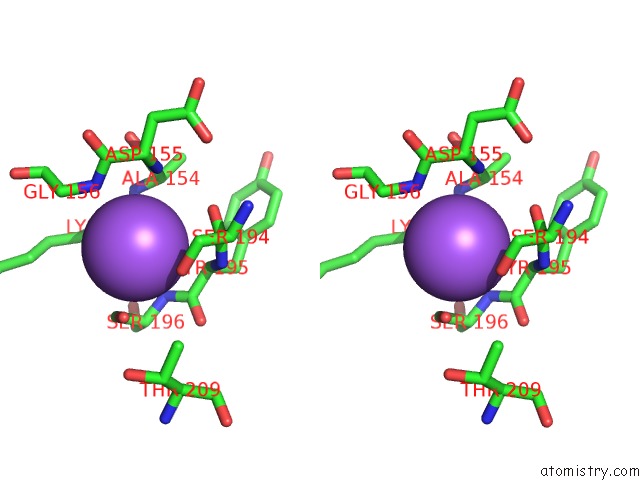
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Crystal Structure of Non-Cardiotoxic Bence-Jones Light Chain Dimer M8 within 5.0Å range:
|
Reference:
L.Oberti,
P.Rognoni,
A.Barbiroli,
F.Lavatelli,
R.Russo,
M.Maritan,
G.Palladini,
M.Bolognesi,
G.Merlini,
S.Ricagno.
Concurrent Structural and Biophysical Traits Link with Immunoglobulin Light Chains Amyloid Propensity. Sci Rep V. 7 16809 2017.
ISSN: ESSN 2045-2322
PubMed: 29196671
DOI: 10.1038/S41598-017-16953-7
Page generated: Mon Oct 7 22:30:27 2024
ISSN: ESSN 2045-2322
PubMed: 29196671
DOI: 10.1038/S41598-017-16953-7
Last articles
Cl in 7VTVCl in 7VRG
Cl in 7VSL
Cl in 7VSI
Cl in 7VQM
Cl in 7VR9
Cl in 7VPE
Cl in 7VRE
Cl in 7VRA
Cl in 7VP8