Sodium »
PDB 5l6q-5lq6 »
5lk3 »
Sodium in PDB 5lk3: Structure of Hantavirus Envelope Glycoprotein Gc in Postfusion Conformation in Presence of 500 Mm Kcl
Protein crystallography data
The structure of Structure of Hantavirus Envelope Glycoprotein Gc in Postfusion Conformation in Presence of 500 Mm Kcl, PDB code: 5lk3
was solved by
P.Guardado-Calvo,
F.A.Rey,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 37.52 / 1.50 |
| Space group | H 3 |
| Cell size a, b, c (Å), α, β, γ (°) | 107.141, 107.141, 127.627, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 17.1 / 20.1 |
Other elements in 5lk3:
The structure of Structure of Hantavirus Envelope Glycoprotein Gc in Postfusion Conformation in Presence of 500 Mm Kcl also contains other interesting chemical elements:
| Potassium | (K) | 1 atom |
Sodium Binding Sites:
The binding sites of Sodium atom in the Structure of Hantavirus Envelope Glycoprotein Gc in Postfusion Conformation in Presence of 500 Mm Kcl
(pdb code 5lk3). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Structure of Hantavirus Envelope Glycoprotein Gc in Postfusion Conformation in Presence of 500 Mm Kcl, PDB code: 5lk3:
In total only one binding site of Sodium was determined in the Structure of Hantavirus Envelope Glycoprotein Gc in Postfusion Conformation in Presence of 500 Mm Kcl, PDB code: 5lk3:
Sodium binding site 1 out of 1 in 5lk3
Go back to
Sodium binding site 1 out
of 1 in the Structure of Hantavirus Envelope Glycoprotein Gc in Postfusion Conformation in Presence of 500 Mm Kcl
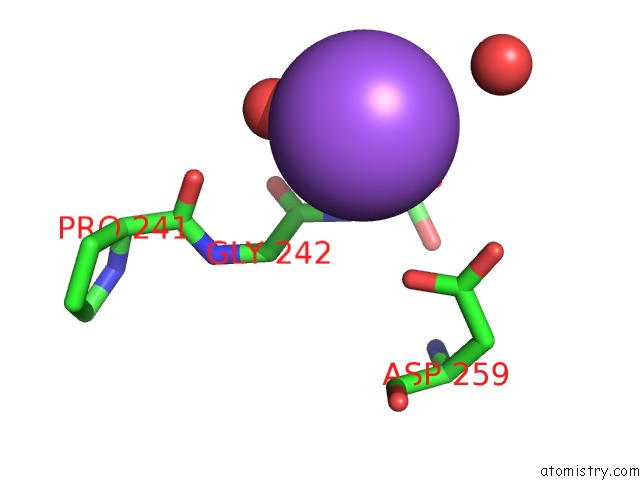
Mono view
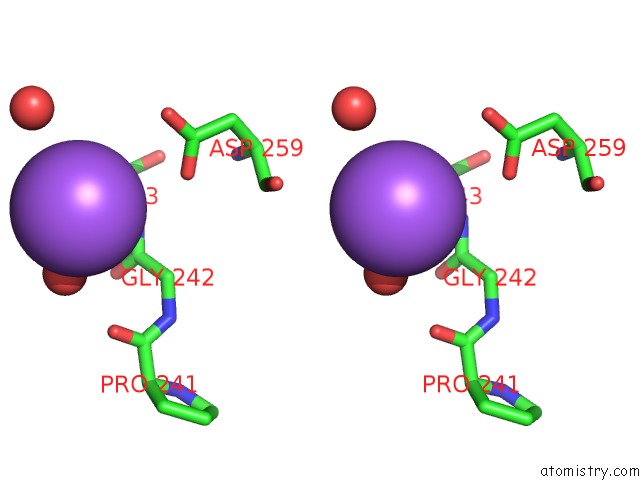
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Structure of Hantavirus Envelope Glycoprotein Gc in Postfusion Conformation in Presence of 500 Mm Kcl within 5.0Å range:
|
Reference:
P.Guardado-Calvo,
E.A.Bignon,
E.Stettner,
S.A.Jeffers,
J.Perez-Vargas,
G.Pehau-Arnaudet,
M.A.Tortorici,
J.L.Jestin,
P.England,
N.D.Tischler,
F.A.Rey.
Mechanistic Insight Into Bunyavirus-Induced Membrane Fusion From Structure-Function Analyses of the Hantavirus Envelope Glycoprotein Gc. Plos Pathog. V. 12 05813 2016.
ISSN: ESSN 1553-7374
PubMed: 27783711
DOI: 10.1371/JOURNAL.PPAT.1005813
Page generated: Mon Oct 7 22:22:20 2024
ISSN: ESSN 1553-7374
PubMed: 27783711
DOI: 10.1371/JOURNAL.PPAT.1005813
Last articles
Zn in 9J0NZn in 9J0O
Zn in 9J0P
Zn in 9FJX
Zn in 9EKB
Zn in 9C0F
Zn in 9CAH
Zn in 9CH0
Zn in 9CH3
Zn in 9CH1