Sodium »
PDB 5kwe-5l6n »
5l0i »
Sodium in PDB 5l0i: Human Metavinculin Mvt R975W Cardiomyopathy-Associated Mutant (Residues 959-1134)
Protein crystallography data
The structure of Human Metavinculin Mvt R975W Cardiomyopathy-Associated Mutant (Residues 959-1134), PDB code: 5l0i
was solved by
K.Chinthalapudi,
T.Izard,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 44.78 / 2.45 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 90.074, 41.519, 44.148, 90.00, 96.14, 90.00 |
| R / Rfree (%) | 19.5 / 23.8 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Human Metavinculin Mvt R975W Cardiomyopathy-Associated Mutant (Residues 959-1134)
(pdb code 5l0i). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total only one binding site of Sodium was determined in the Human Metavinculin Mvt R975W Cardiomyopathy-Associated Mutant (Residues 959-1134), PDB code: 5l0i:
In total only one binding site of Sodium was determined in the Human Metavinculin Mvt R975W Cardiomyopathy-Associated Mutant (Residues 959-1134), PDB code: 5l0i:
Sodium binding site 1 out of 1 in 5l0i
Go back to
Sodium binding site 1 out
of 1 in the Human Metavinculin Mvt R975W Cardiomyopathy-Associated Mutant (Residues 959-1134)
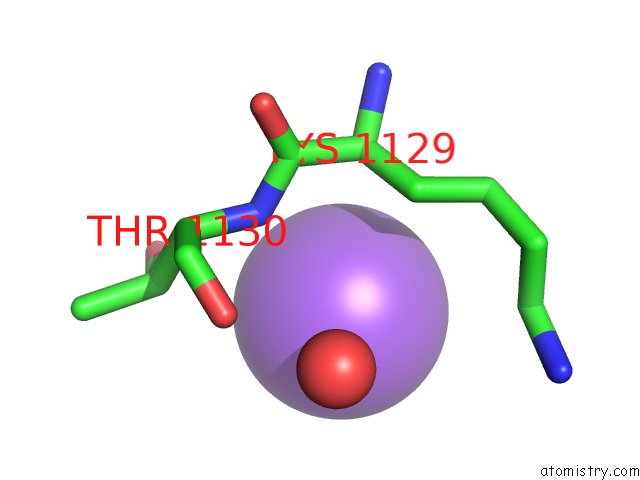
Mono view
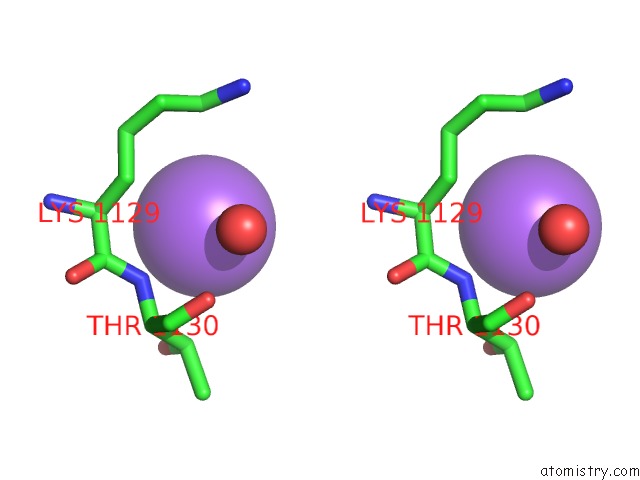
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Human Metavinculin Mvt R975W Cardiomyopathy-Associated Mutant (Residues 959-1134) within 5.0Å range:
|
Reference:
K.Chinthalapudi,
E.S.Rangarajan,
D.T.Brown,
T.Izard.
Differential Lipid Binding of Vinculin Isoforms Promotes Quasi-Equivalent Dimerization. Proc.Natl.Acad.Sci.Usa V. 113 9539 2016.
ISSN: ESSN 1091-6490
PubMed: 27503891
DOI: 10.1073/PNAS.1600702113
Page generated: Mon Aug 18 00:46:54 2025
ISSN: ESSN 1091-6490
PubMed: 27503891
DOI: 10.1073/PNAS.1600702113
Last articles
Na in 7E4CNa in 7E21
Na in 7E1Z
Na in 7E4A
Na in 7E02
Na in 7DTF
Na in 7DTB
Na in 7DPH
Na in 7DPD
Na in 7DNB