Sodium »
PDB 5g3g-5gwa »
5glk »
Sodium in PDB 5glk: Crystal Structure of COXYL43, GH43 Beta-Xylosidase/Alpha- Arabinofuranosidase From A Compost Microbial Metagenome, Calcium-Free Form.
Protein crystallography data
The structure of Crystal Structure of COXYL43, GH43 Beta-Xylosidase/Alpha- Arabinofuranosidase From A Compost Microbial Metagenome, Calcium-Free Form., PDB code: 5glk
was solved by
T.Matsuzawa,
N.Kishine,
Z.Fujimoto,
K.Yaoi,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 78.87 / 1.70 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 74.483, 61.327, 79.226, 90.00, 95.44, 90.00 |
| R / Rfree (%) | 15.8 / 18.6 |
Sodium Binding Sites:
The binding sites of Sodium atom in the Crystal Structure of COXYL43, GH43 Beta-Xylosidase/Alpha- Arabinofuranosidase From A Compost Microbial Metagenome, Calcium-Free Form.
(pdb code 5glk). This binding sites where shown within
5.0 Angstroms radius around Sodium atom.
In total 4 binding sites of Sodium where determined in the Crystal Structure of COXYL43, GH43 Beta-Xylosidase/Alpha- Arabinofuranosidase From A Compost Microbial Metagenome, Calcium-Free Form., PDB code: 5glk:
Jump to Sodium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Sodium where determined in the Crystal Structure of COXYL43, GH43 Beta-Xylosidase/Alpha- Arabinofuranosidase From A Compost Microbial Metagenome, Calcium-Free Form., PDB code: 5glk:
Jump to Sodium binding site number: 1; 2; 3; 4;
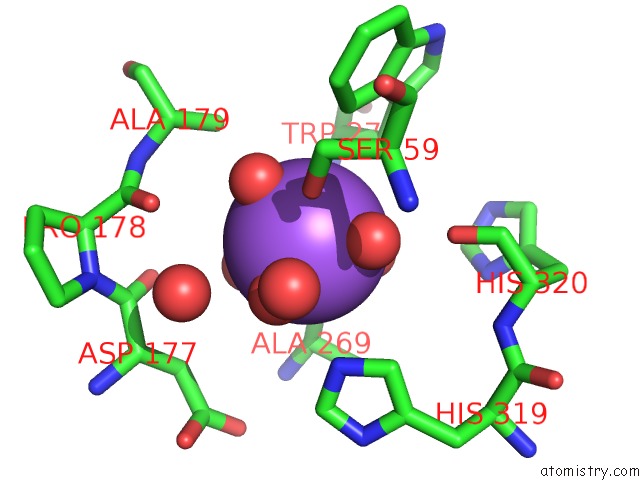
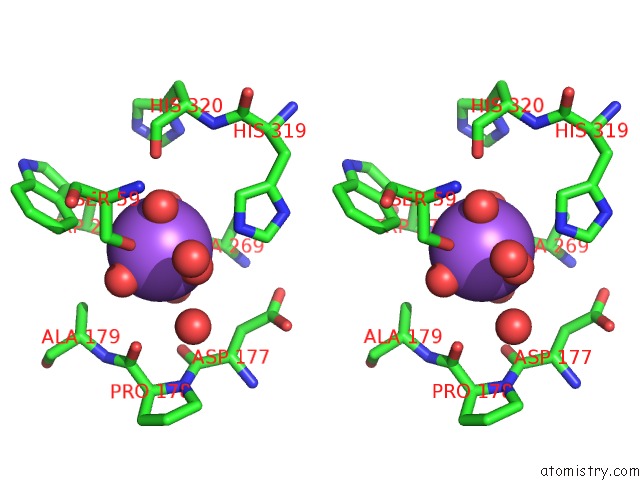
Sodium binding site 1 out of 4 in 5glk
Go back to
Sodium binding site 1 out
of 4 in the Crystal Structure of COXYL43, GH43 Beta-Xylosidase/Alpha- Arabinofuranosidase From A Compost Microbial Metagenome, Calcium-Free Form.
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 1 of Crystal Structure of COXYL43, GH43 Beta-Xylosidase/Alpha- Arabinofuranosidase From A Compost Microbial Metagenome, Calcium-Free Form. within 5.0Å range:
|
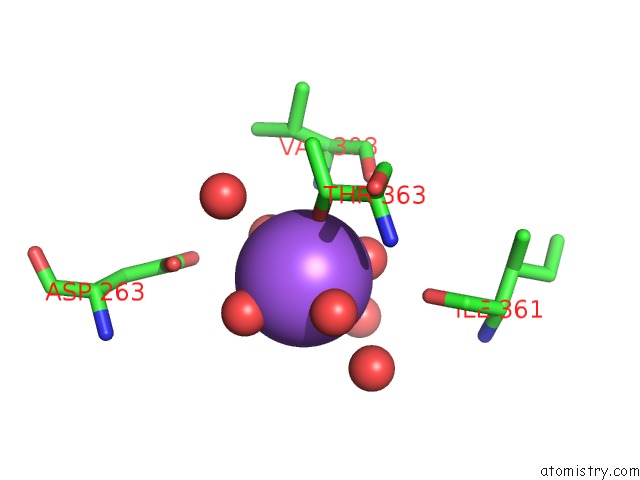
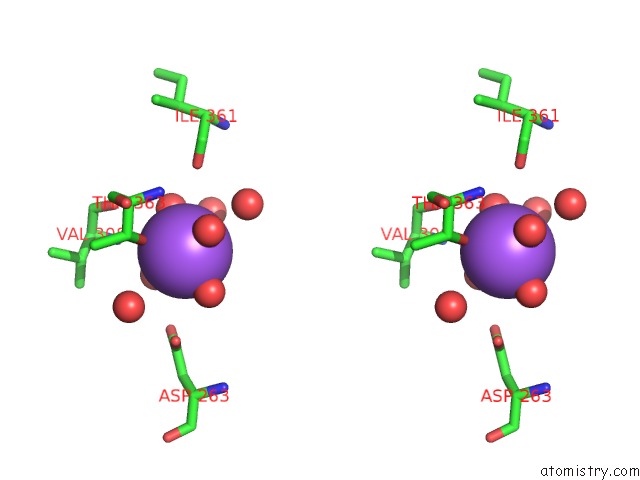
Sodium binding site 2 out of 4 in 5glk
Go back to
Sodium binding site 2 out
of 4 in the Crystal Structure of COXYL43, GH43 Beta-Xylosidase/Alpha- Arabinofuranosidase From A Compost Microbial Metagenome, Calcium-Free Form.
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 2 of Crystal Structure of COXYL43, GH43 Beta-Xylosidase/Alpha- Arabinofuranosidase From A Compost Microbial Metagenome, Calcium-Free Form. within 5.0Å range:
|
Sodium binding site 3 out of 4 in 5glk
Go back to
Sodium binding site 3 out
of 4 in the Crystal Structure of COXYL43, GH43 Beta-Xylosidase/Alpha- Arabinofuranosidase From A Compost Microbial Metagenome, Calcium-Free Form.
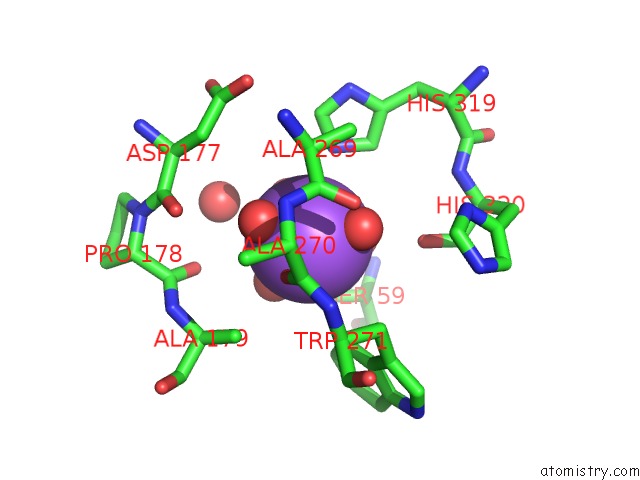
Mono view
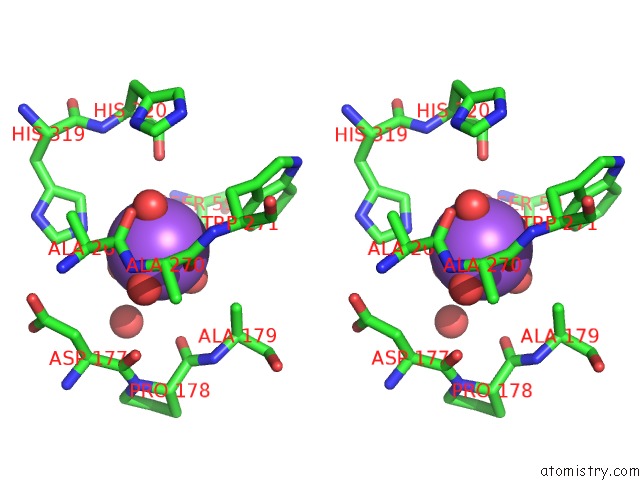
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 3 of Crystal Structure of COXYL43, GH43 Beta-Xylosidase/Alpha- Arabinofuranosidase From A Compost Microbial Metagenome, Calcium-Free Form. within 5.0Å range:
|
Sodium binding site 4 out of 4 in 5glk
Go back to
Sodium binding site 4 out
of 4 in the Crystal Structure of COXYL43, GH43 Beta-Xylosidase/Alpha- Arabinofuranosidase From A Compost Microbial Metagenome, Calcium-Free Form.
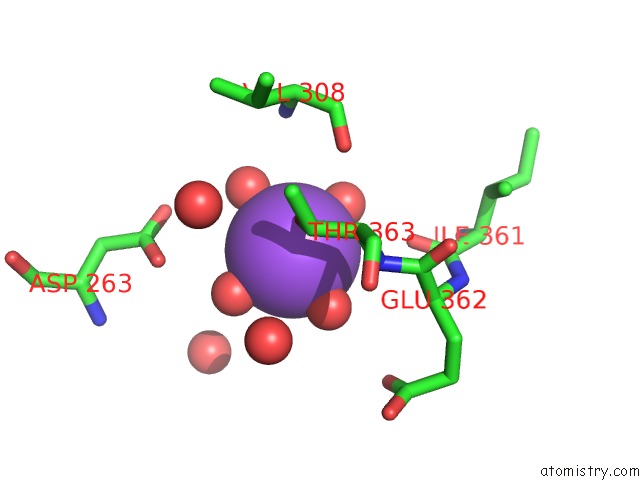
Mono view
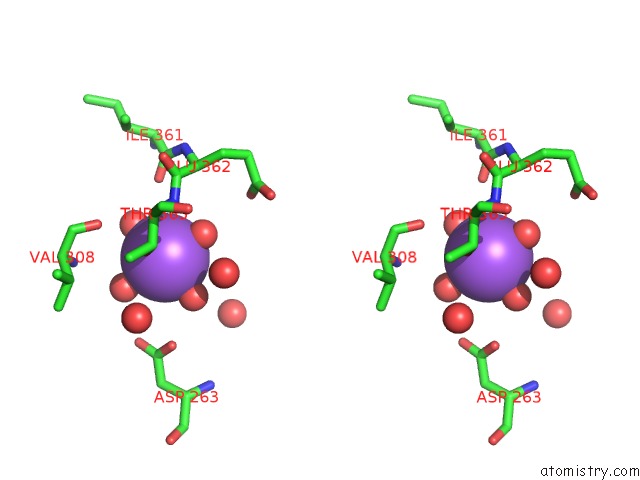
Stereo pair view

Mono view

Stereo pair view
A full contact list of Sodium with other atoms in the Na binding
site number 4 of Crystal Structure of COXYL43, GH43 Beta-Xylosidase/Alpha- Arabinofuranosidase From A Compost Microbial Metagenome, Calcium-Free Form. within 5.0Å range:
|
Reference:
T.Matsuzawa,
S.Kaneko,
N.Kishine,
Z.Fujimoto,
K.Yaoi.
Crystal Structure of Metagenomic Beta-Xylosidase/ Alpha-L-Arabinofuranosidase Activated By Calcium. J. Biochem. V. 162 173 2017.
ISSN: ISSN 1756-2651
PubMed: 28204531
DOI: 10.1093/JB/MVX012
Page generated: Mon Oct 7 21:12:47 2024
ISSN: ISSN 1756-2651
PubMed: 28204531
DOI: 10.1093/JB/MVX012
Last articles
F in 4CFNF in 4CLJ
F in 4CLI
F in 4CK7
F in 4CK6
F in 4CKA
F in 4CGN
F in 4C7O
F in 4CFT
F in 4CEX